-
Notifications
You must be signed in to change notification settings - Fork 5
Home
Ultraliser is an unconditionally robust and optimized framework dedicated primarily to in silico neuroscience research. Ultraliser is capable of generating high fidelity and multiscale 3D neuroscientific models - such as: nuclei, mitochondria, endoplasmic reticula, neurons, astrocytes, pericytes, neuronal branches with dendritic spines, minicolumns with thousands of neurons and large networks of cerebral vasculature - with realistic geometries.
Ultraliser implements an effective voxelization-based remeshing engine that can rasterize non-watertight surface meshes - in the form of triangular soups - into high resolution volumes, with which we can reconstruct topologically accurate, adaptively optimized and watertight surface manifolds.
In addition to their importance for accurate quantitative analysis, resulting models are primarily intended to automate the process of conducting supercomputer simulations of neuroscience experiments; complementing in vivo and in vitro techniques.
Watertight triangular meshes are used for (i) performing 3D particle simulations, (ii) mesh-based skeletonization, in which accurate morphologies of cellular structures are obtained for performing 1D compartmental simulations and (iii) tetrahedralization, in which we can generate tetrahedral volume meshes for 3D reaction-diffusion simulations. Annotated volumetric tissue models are also used in in silico imaging studies, where we can simulate optical imaging experiments with brightfield or fluorescence microscopy10. A high-level overview of Ultraliser's workflow is graphically illustrated in Figure 1.
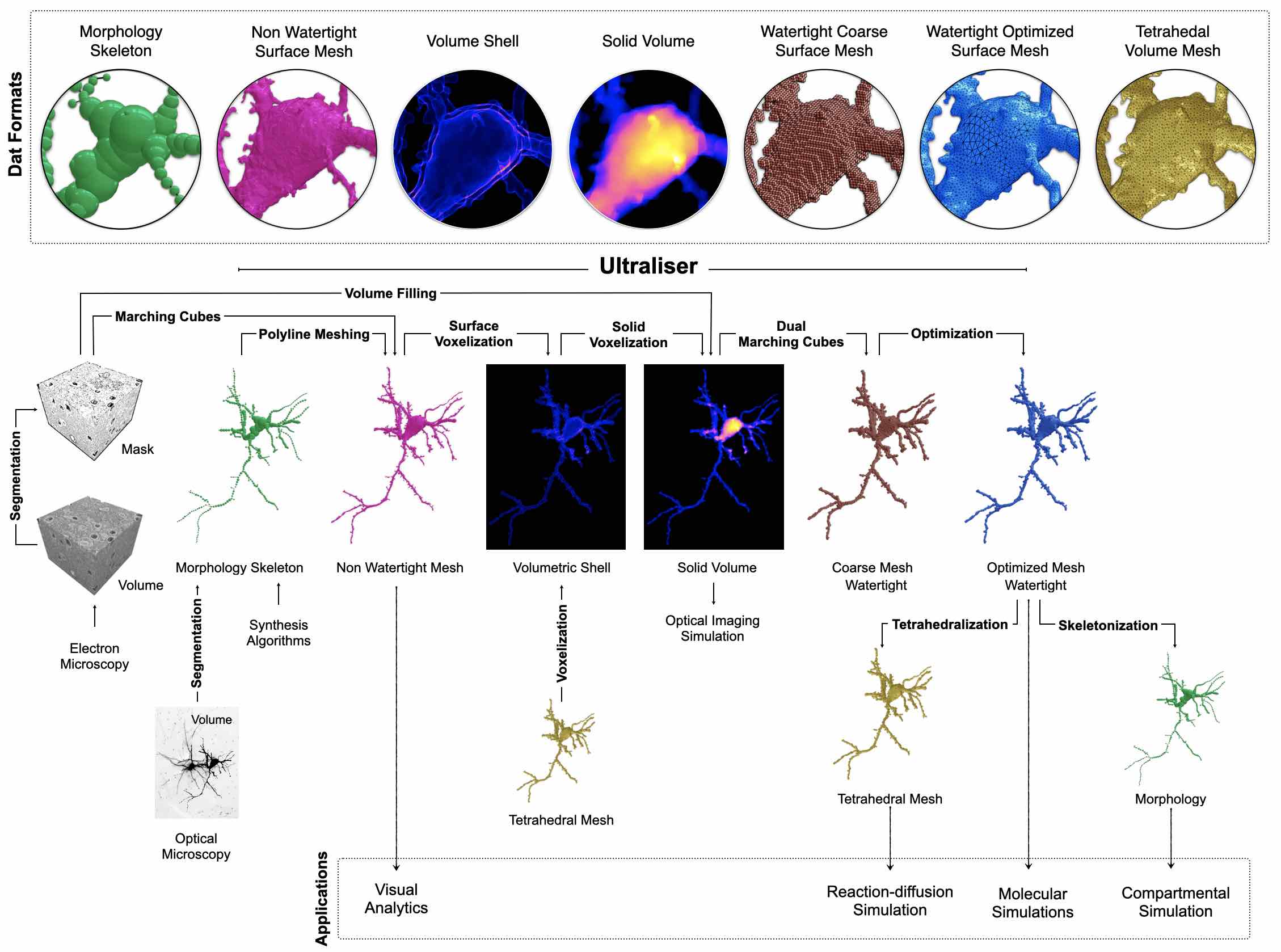
Figure 1 A high-level overview of Ultraliser's workflow showing its subsequent stages (surface and solid voxelization, mesh reconstruction and optimization) including the different data formats that can be processed within its workflow (masks, gray-scale volumes, polygonal surface meshes, morphological skeletons and tetrahedral volumetric meshes). Ultraliser creates high-fidelity, adaptively optimized and watertight surface manifolds and large-scale annotated volumes. The surface meshes are used directly for molecular simulations, and can be further processed to create volumetric (tetrahedral or hexahedral) meshes for performing reaction-diffusion simulation. They can be also used by mesh-based skeletonization applications to skeletonize accurate morphological counterparts. The annotated volumes are used in in silico optical imaging experiments10.
- Reconstruction of high fidelity, optimized1 and two-manifold watertight2 triangular mesh models from non-watertight inputs represented by polygonal soups.
- Surface mesh smoothing and optimization using Laplacian operators and feature-preserving adaptive mesh optimization1.
- Reconstruction of large-scale volumetric models3 from non-watertight input meshes using high performance surface and solid voxelization.
- Reconstruction of optimized and smooth surface meshes from input volumes using parallel implementations of the standard marching cubes4 algorithm and the advanced dual marching cubes5 algorithm.
- Reconstruction of optimized and smooth surface meshes from input binary masks of segmened data.
- Reconstruction of geometrically realistic watertight mesh models of spiny neurons from corresponding morphological skeletons6 .
- Reconstruction of geometrically realistic watertight mesh models of complete astroglial cells8 (with endfeet) from input morphological skeletons and endfeet surface patches9 .
- Reconstruction of high-fidelity, optimized and multi-partitioned vascular meshes from fragmented and large-scale vascular network graphs7.
- Morphology, mesh and volume quantiative and qualitative analysis.
- Generation of color-coded multi-axis projections of spatial data (morphologies, meshes and volumes) for visual analytics.
Ultraliser uses the CMake build system to compile. Detailed installation instructions are available in this page.
Ultraliser comes integrated with the following primary applications:
-
ultraMesh2Mesh, which reconstructs a two-manifold watertight mesh from a non-watertight input mesh. (Documentation, Examples)
-
ultraMeshes2Mesh, which reconstructs a two-manifold watertight mesh from a list of non-watertight input meshes (Documentation, Examples)
-
ultraMesh2Volume, which reconstructs a large-scale tagged volume from a non -watertight input mesh. (Documentation, Examples)
-
ultraVolume2Mesh, which reconstructs a two-manifold watertight mesh from an input volume, based on a given isovalue. (Documentation, Examples)
-
ultraMask2Mesh, which reconstructs a two-manifold watertight mesh from a segmented binary mask in .TIFF format. (Documentation, Examples)
-
ultraNeuroMorpho2Mesh, which reconstructs a two-manifold watertight neuronal surface mesh from its morphological description. (Documentation, Examples)
-
ultraAstroMorpho2Mesh, which reconstructs a two-manifold watertight astroglial surface mesh from its morphological description including endfeet data. (Documentation, Examples)
-
ultraVessMorpho2Mesh, which reconstructs a two-manifold watertight vascular surface mesh from an input graph that represents the vascular network. (Documentation, Examples)
Ultraliser integrates a list of other secondary applications including:
-
ultraH5MeshConverter, which converts an input mesh stored in H5 format (the format is available from the MICrONS Explorer) into .OBJ, .PLY, .OFF or .STL standard format. (Documentation)
-
ultraMeshQualityChecker
-
ultraCleanMesh
-
ultraSampleVascularMorphology
-
ultraScaleMesh, which scales an input mesh along the XYZ dimensions -efficiently- based on a user-defined scale factor. This application is required when the measurements of the model are made at nm scale, not at um scale.
-
ultraSplitPartitions, which splits an input mesh with multiple partitions into multiple output meshes, each has a single partition (or island). This application checks also if the input mesh is watertight or not. (Documentation)
-
ultraVerifyVMVConnectivity, which efficiently verifies the connectivity of an input VMV vascular network to ensure that the entire graph is connected. If the input graph does not have the section-to-section connectivity information, this application creates a new graph that includes the section-to-section connectivity data. (Documentation)
Please refer to the github issue tracker for fixed and open bugs. User can also report any bugs and request new features needed for their research. We are happy to provide direct support .
Ultraliser is available to download and use under the GNU General Public License, version 3 (GPL, or “free software”). The code is open sourced with approval from the open sourcing committee and principal coordinators of the Blue Brain Project in March 2021.
The volume reconstruction algorithms in Ultraliser are based on the following paper.
@article{abdellah2017reconstruction,
title={Reconstruction and visualization of large-scale volumetric models of neocortical
circuits for physically-plausible in silico optical studies},
author={Abdellah, Marwan and Hernando, Juan and Antille, Nicolas and Eilemann, Stefan and
Markram, Henry and Sch{\"u}rmann, Felix},
journal={BMC bioinformatics},
volume={18},
number={10},
pages={402},
year={2017},
publisher={BioMed Central}
}
Ultraliser is developed by the Visualization team at the Blue Brain Project, Ecole Polytechnique Federale de Lausanne (EPFL). Financial support was provided by competitive research funding from King Abdullah University of Science and Technology (KAUST).
-
The volume reconstruction code is an extension to the work of Marwan Abdellah's PhD (In silico Brain Imaging: Physically-plausible Methods for Visualizing Neocortical Microcircuitry).
-
The mesh optimization code in Ultraliser is based on the routiens provided by the GAMer (Geometry-preserving Adaptive MeshER) library. GAMer is developed by Z. Yu, M. Holst, Y. Cheng, and J.A. McCammon, and can be redistributed and modified under the terms of the GNU General Public License as published by the Free Software Foundation; either version 2 of the License, or any later version.
-
The watertighness verification code in Ultraliser is based on an extended version of the MeshFix library. MeshFix is developed by Marco Attene, Consiglio Nazionale delle Ricerche, Istituto di Matematica Applicata e Tecnologie Informatiche, Sezione di Genova, IMATI-GE / CNR, and can be redistributed and modified under the terms of the GNU General Public License as published by the Free Software Foundation; either version 3 of the License, or any later version.
-
The mesh analysis code is implemented based on the metrics described in the The Verdict Geometric Quality Library.
-
The morphology analysis code is implemented based on the metrics described in NeuroMorphoVis.
-
The values of the colormaps used to generate the color-coded projections are obtained from the matplotlib library.
-
The H5 morphology samples are available with permissions from the Blue Brain Project, Ecole Polytechnique Federale de Lausanne (EPFL).
-
The SWC morphology samples of neurons and astrocytes are available from NeuroMorpho.Org. NeuroMorpho.Org is a centrally curated inventory or repository of digitally reconstructed neurons associated with peer-reviewed publications.
-
The SWC morphology samples of brain vasculature are available from the Brain Vasculature (BraVa) database. The Brain Vasculature (BraVa) database contains digital reconstructions of the human brain arterial arborizations from 61 healthy adult subjects along with extracted morphological measurements. The arterial arborizations include the six major trees stemming from the circle of Willis, namely: the left and right Anterior Cerebral Arteries (ACAs), Middle Cerebral Arteries (MCAs), and Posterior Cerebral Arteries (PCAs). Citation: Susan N. Wright, Peter Kochunov, Fernando Mut Maurizio Bergamino, Kerry M. Brown, John C. Mazziotta, Arthur W. Toga, Juan R. Cebral, Giorgio A. Ascoli. Digital reconstruction and morphometric analysis of human brain arterial vasculature from magnetic resonance angiography. NeuroImage, 82, 170-181, (2013).
-
The VMV vascular morphologies are available with permissions from Pablo Blinder, Department of Neurobiology, Faculty of Life Sciences at Tel Aviv University.
-
The H5 vascular morphologies are available with permissions from the Blue Brain Project, Ecole Polytechnique Federale de Lausanne (EPFL). The original dataset courtesy of Bruno Weber, University of Zurich, Switzerland.
-
The table of contents for all the user documentation pages are generated with markdown-toc.
For more information on Ultraliser, comments or suggestions, please contact:
Marwan Abdellah
Scientific Visualiation Engineer
Blue Brain Project
marwan.abdellah@epfl.ch
Felix Schürmann
Co-director of the Blue Brain Project
felix.schuermann@epfl.ch
Should you have any questions concerning press enquiriries, please contact:
Kate Mullins
Communications
Blue Brain Project
kate.mullins@epfl.ch
-
YU, Zeyun, HOLST, Michael J., CHENG, Yuhui, et al. Feature-preserving adaptive mesh generation for molecular shape modeling and simulation. Journal of Molecular Graphics and Modelling, 2008, vol. 26, no 8, p. 1370-1380.
-
ATTENE, Marco. A lightweight approach to repairing digitized polygon meshes. The visual computer, 2010, vol. 26, no 11, p. 1393-1406.
-
ABDELLAH, Marwan, HERNANDO, Juan, ANTILLE, Nicolas, et al. Reconstruction and visualization of large-scale volumetric models of neocortical circuits for physically-plausible in silico optical studies. BMC bioinformatics, 2017, vol. 18, no 10, p. 39-50.
-
LORENSEN, William E. et CLINE, Harvey E. Marching cubes: A high resolution 3D surface construction algorithm. ACM siggraph computer graphics, 1987, vol. 21, no 4, p. 163-169.
-
NIELSON, Gregory M. Dual marching cubes. In : IEEE visualization 2004. IEEE, 2004. p. 489-496.
-
ABDELLAH, Marwan, HERNANDO, Juan, EILEMANN, Stefan, et al. NeuroMorphoVis: a collaborative framework for analysis and visualization of neuronal morphology skeletons reconstructed from microscopy stacks. Bioinformatics, 2018, vol. 34, no 13, p. i574-i582.
-
ABDELLAH, Marwan, GUERRERO, Nadir Román, LAPERE, Samuel, et al. Interactive visualization and analysis of morphological skeletons of brain vasculature networks with VessMorphoVis. Bioinformatics, 2020, vol. 36, no Supplement_1, p. i534-i541.
-
ZISIS, Eleftherios, KELLER, Daniel, KANARI, Lida, et al. Digital reconstruction of the neuro-glia-vascular architecture. Cerebral Cortex, 2021, vol. 31, no 12, p. 5686-5703.
-
ABDELLAH, Marwan, FONI, Alessandro, ZISIS, Eleftherios, et al. Metaball skinning of synthetic astroglial morphologies into realistic mesh models for in silico simulations and visual analytics. Bioinformatics, 2021, vol. 37, no Supplement_1, p. i426-i433.
-
ABDELLAH, Marwan. In silico brain imaging: physically-plausible methods for visualizing neocortical microcircuitry. EPFL, 2017.
Copyright © 2022 - Blue Brain Project / Ecole Polytechnique Fédérale de Lausanne (EPFL)
