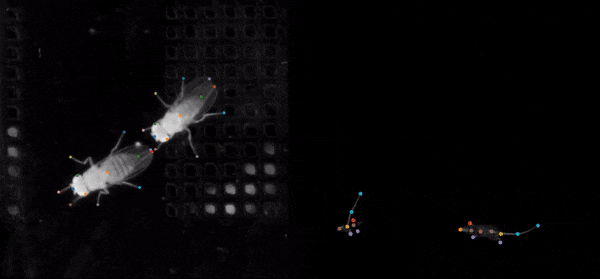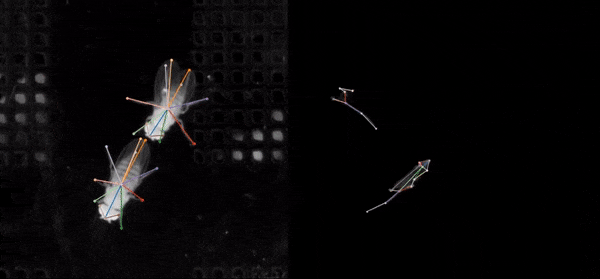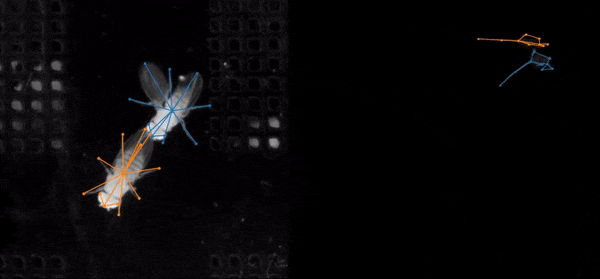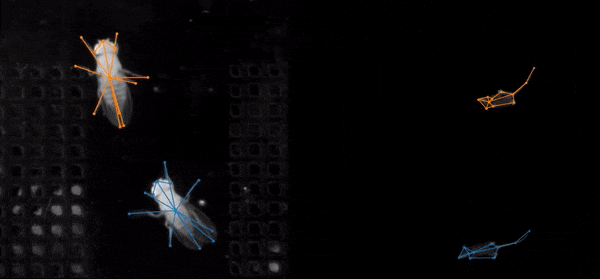
SLEAP is an open source deep-learning based framework for multi-animal pose tracking (Pereira et al., Nature Methods, 2022). It can be used to track any type or number of animals and includes an advanced labeling/training GUI for active learning and proofreading.
- Easy, one-line installation with support for all OSes
- Purpose-built GUI and human-in-the-loop workflow for rapidly labeling large datasets
- Single- and multi-animal pose estimation with top-down and bottom-up training strategies
- State-of-the-art pretrained and customizable neural network architectures that deliver accurate predictions with very few labels
- Fast training: 15 to 60 mins on a single GPU for a typical dataset
- Fast inference: up to 600+ FPS for batch, <10ms latency for realtime
- Support for remote training/inference workflow (for using SLEAP without GPUs)
- Flexible developer API for building integrated apps and customization
SLEAP is installed as a Python package. We strongly recommend using Miniconda to install SLEAP in its own environment.
You can find the latest version of SLEAP in the Releases page.
conda (Windows/Linux/GPU):
conda create -y -n sleap -c conda-forge -c nvidia -c sleap -c anaconda sleappip (any OS except Apple silicon):
pip install sleap[pypi]See the docs for full installation instructions.
- Learn step-by-step: Tutorial
- Learn more advanced usage: Guides and Notebooks
- Learn by watching: ABL:AOC 2023 Workshop and MIT CBMM Tutorial
- Learn by reading: Paper (Pereira et al., Nature Methods, 2022) and Review on behavioral quantification (Pereira et al., Nature Neuroscience, 2020)
- Learn from others: Discussions on Github
SLEAP is the successor to the single-animal pose estimation software LEAP (Pereira et al., Nature Methods, 2019).
If you use SLEAP in your research, please cite:
T.D. Pereira, N. Tabris, A. Matsliah, D. M. Turner, J. Li, S. Ravindranath, E. S. Papadoyannis, E. Normand, D. S. Deutsch, Z. Y. Wang, G. C. McKenzie-Smith, C. C. Mitelut, M. D. Castro, J. D’Uva, M. Kislin, D. H. Sanes, S. D. Kocher, S. S-H, A. L. Falkner, J. W. Shaevitz, and M. Murthy. Sleap: A deep learning system for multi-animal pose tracking. Nature Methods, 19(4), 2022
BibTeX:
@ARTICLE{Pereira2022sleap,
title={SLEAP: A deep learning system for multi-animal pose tracking},
author={Pereira, Talmo D and
Tabris, Nathaniel and
Matsliah, Arie and
Turner, David M and
Li, Junyu and
Ravindranath, Shruthi and
Papadoyannis, Eleni S and
Normand, Edna and
Deutsch, David S and
Wang, Z. Yan and
McKenzie-Smith, Grace C and
Mitelut, Catalin C and
Castro, Marielisa Diez and
D'Uva, John and
Kislin, Mikhail and
Sanes, Dan H and
Kocher, Sarah D and
Samuel S-H and
Falkner, Annegret L and
Shaevitz, Joshua W and
Murthy, Mala},
journal={Nature Methods},
volume={19},
number={4},
year={2022},
publisher={Nature Publishing Group}
}
}
Follow @talmop on Twitter for news and updates!
Technical issue with the software?
- Check the Help page.
- Ask the community via discussions on Github.
- Search the issues on GitHub or open a new one.
General inquiries? Reach out to talmo@salk.edu.
- Talmo Pereira, Salk Institute for Biological Studies
- Liezl Maree, Salk Institute for Biological Studies
- Arlo Sheridan, Salk Institute for Biological Studies
- Arie Matsliah, Princeton Neuroscience Institute, Princeton University
- Nat Tabris, Princeton Neuroscience Institute, Princeton University
- David Turner, Research Computing and Princeton Neuroscience Institute, Princeton University
- Joshua Shaevitz, Physics and Lewis-Sigler Institute, Princeton University
- Mala Murthy, Princeton Neuroscience Institute, Princeton University
SLEAP was created in the Murthy and Shaevitz labs at the Princeton Neuroscience Institute at Princeton University.
SLEAP is currently being developed and maintained in the Talmo Lab at the Salk Institute for Biological Studies, in collaboration with the Murthy and Shaevitz labs at Princeton University.
This work was made possible through our funding sources, including:
- NIH BRAIN Initiative R01 NS104899
- Princeton Innovation Accelerator Fund
SLEAP is released under a Clear BSD License and is intended for research/academic use only. For commercial use, please contact: Laurie Tzodikov (Assistant Director, Office of Technology Licensing), Princeton University, 609-258-7256.