This is the repository for Shiny Contest submission. This repository contains code for the default deployment of systemPipeShiny.
To deploy the same app, first install systemPipeShiny.
Then, it should be ready to go. If you need to change some app options and settings,
all you need is to modify the global.R file and read our manual
for details.
systemPipeShiny (SPS) is an R/Bioconductor package that provides a Shiny application for interactive data analysis workflow management and downstream data visualization. SPS allows non-R users, such as experimentalists, to design, control, execute and visualize data analysis workflows interactively without requiring knowledge of R. Moreover, SPS has been designed as a general purpose framework to allow users to perform deep customization and add their own components.
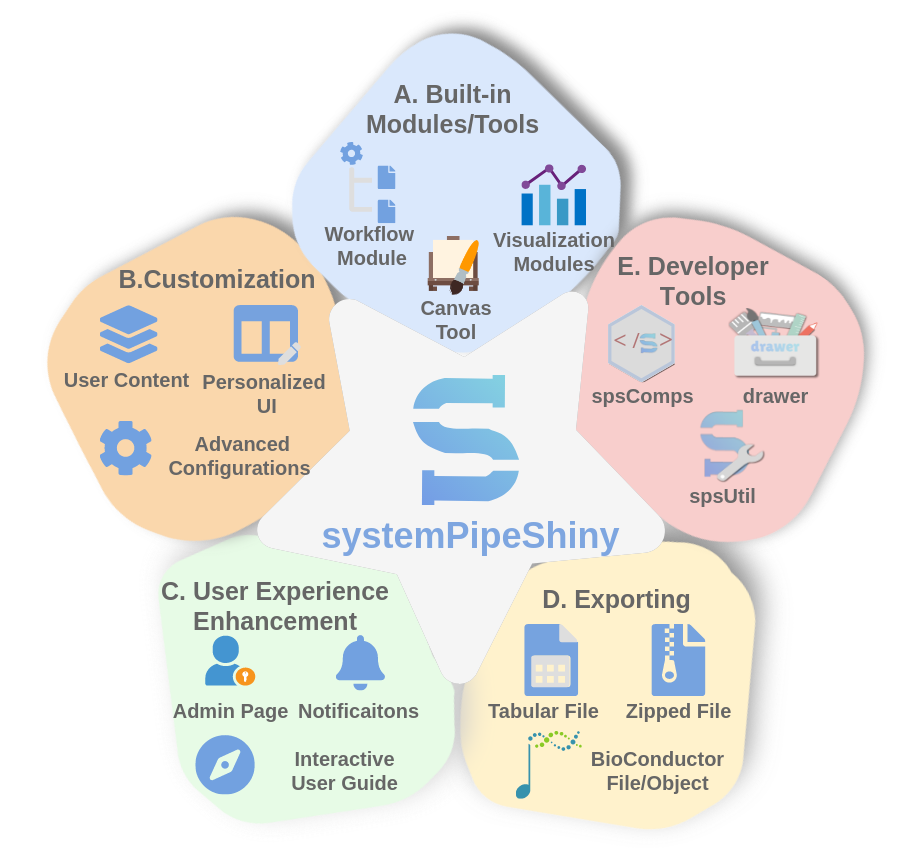
Figure 1. The overview of SPS
SPS main app is designed in a dashboard UI with default modules, user-defined tabs and the Canvas as individual dashboard tabs. There is also another dashboard, the admin page, where the app maintainer can manage the app credentials and see app information interactively. Besides, SPS provides enriched supporting packages as R/Shiny extensions for users to use on their own projects if they do not want to use SPS framework.
- Three powerful default modules:
- Workflow: use systemPipeR (SPR) as backend to interactively generate, design, and run workflows. Current templates are biological-centric, but as a workflow environment, SPR can adapt to any type of data analysis workflow.
- RNAseq: Interactive conduct downstream analysis of RNA sequencing raw counts, including different ways of data normalization, deferentially expressed gene analysis, and more than 10 types of clustering or summary plotting methods.
- Quick ggplot: To make a ggplot within a few mouse clicks with any kind of tabular data you upload.
- More modules in the future...
- Extendable: users are able to add their own components to the dashboard as individual "tabs".
- Different templates that satisfy both beginners and advanced developers
- Simple: high-level wrappers, you only need to focus on plotting code, no Shiny development knowledge is required.
- Full: full Shiny code, for advanced users, customize everything of the tab.
- Modularization: the whole app is built on top of Shiny modules, which gives it the flexibility to add new content without concerning conflicting with any existing content. Each "tab" on the dashboard is isolated with its own environment.
- Different templates that satisfy both beginners and advanced developers
- Canvas: A workbench for interactive quick image editing
- Communicates with all plot options on other tabs, take screenshots of them and send to this tool for further image editing.
- Combine/compare different plots, add annotations and make a scientific figure.
- Drag to upload your own images.
- Fully customizable: change almost everything of the default app
- Security and admin:
- Options to turn on login feature with interactive loading screens that users can play with
- User-defined secret URL for the admin page
- View real-time app analytics
- Control app accounts interactively
- Notification: a simple way to broadcast messages to your users without redeploying the app every time.
- User defined interactive tutorials: Easy-to-user methods and templates to create your own interactive tutorials
- Detailed debugging: options to turn on verbose logging and traceback messages.
- Duel-end logging: Exceptions handling with logging on both user-end and server-end.
- Deploy-ready: When you initialize an SPS project, the app is deploy-ready. You can deploy it to Shiny servers like shinyapps.io as soon as the project is created.
- Developer tools: If you see any good features from SPS, you can use them in your own apps. Core features of SPS have been split into smaller supporting packages {spsComps}, {drawer} and {spsUtil} for you to use in your own Shiny apps or Rmarkdowns.
Current examples, templates, workflows, and visualizations are biological/bioinformatic focused. However, with the proper configuration, one can apply SPS as a framework for any type of data analysis and visualization.
The submission demo link is only a small part of what SPS can do (defaults). To view how the app differs under different options, try these additional demos:
| Demos | option changed | notes |
|---|---|---|
| Default full installation | See installation | default full app, may take longer (~15s) to load |
| Minimum installation | See installation | no modules installed |
| Login enabled | login_screen = TRUE; login_theme = "empty" |
no modules installed |
| Login and login themes | login_screen = TRUE; login_theme = "random" |
no modules installed |
| App admin page | admin_page = TRUE |
use the link or simply add "?admin" to the end of URL of any demos |
For the login required demos, the app account name is "user" password "user".
For the admin login, account name "admin", password "admin".
Please DO NOT delete or change the password when you are trying the admin features. Although shinyapps.io will reset the app once a while, this will affect other people who are viewing the demo simultaneously.
Conduct our Github README or website for details.
SPS is under heavy development, we recommend you to install the Github version for the latest features.
The RStudio Cloud project associated with this submission does not have Workflow and RNAseq module installed. These two modules depend heavily on some Bioconductor packages that require more than 1GB of RAM to install and to load, which is not available for a typical free account. If you have an advanced account and want to try these two modules. Follow the app start-up message to install packages and restart the app to load the modules.
| Package | Description | Documents | Function reference | Demo |
|---|---|---|---|---|
| SPS main package | website | link | demo | |
| SPS UI and server components | website | link | demo | |
| SPS interactive image editing tool | website | link | demo | |
| SPS utility functions | website | link | NA |
Le Zhang, Daniela Cassol, Ponmathi Ramasamy, Jianhai Zhang, Gordon Mosher & Thomas Girke