Omnipy is a high level Python library for type-driven data wrangling and scalable workflow orchestration.
- June 22, 2024: We're not very good at writing updates. Expect a larger update soon on an important
and potentially groundbreaking new feature of Omnipy: the capability of model objects to automatically
mimic behaviour of the modelled class – with the addition of snapshots and rollbacks.
So e.g.
Model[list[int]]()is not just a run-time typesafe parser that continuously makes sure that the elements in the list are, in fact, integers; the object can also be operated as a list using e.g..append(),.insert()and concatenation with the+operator; and furthermore: if you append an unparseable element, say"abc"instead of"123", it will roll back the contents to the previously validated snapshot! - Feb 3, 2023: Documentation of the Omnipy API is still sparse. However, for examples of running code, please check out the omnipy-examples repo.
- Dec 22, 2022: Omnipy is the new name of the Python package formerly known as uniFAIR. We are very grateful to Dr. Jamin Chen, who gracefully transferred ownership of the (mostly unused) "omnipy" name in PyPI to us!_
(NOTE: Read the section Transformation on the FAIRtracks.net website for a more detailed and better formatted version of the following description!)
Omnipy is designed primarily to simplify development and deployment of (meta)data transformation processes in the context of FAIRification and data brokering efforts. However, the functionality is very generic and can also be used to support research data (and metadata) transformations in a range of fields and contexts beyond life science, including day-to-day research scenarios:
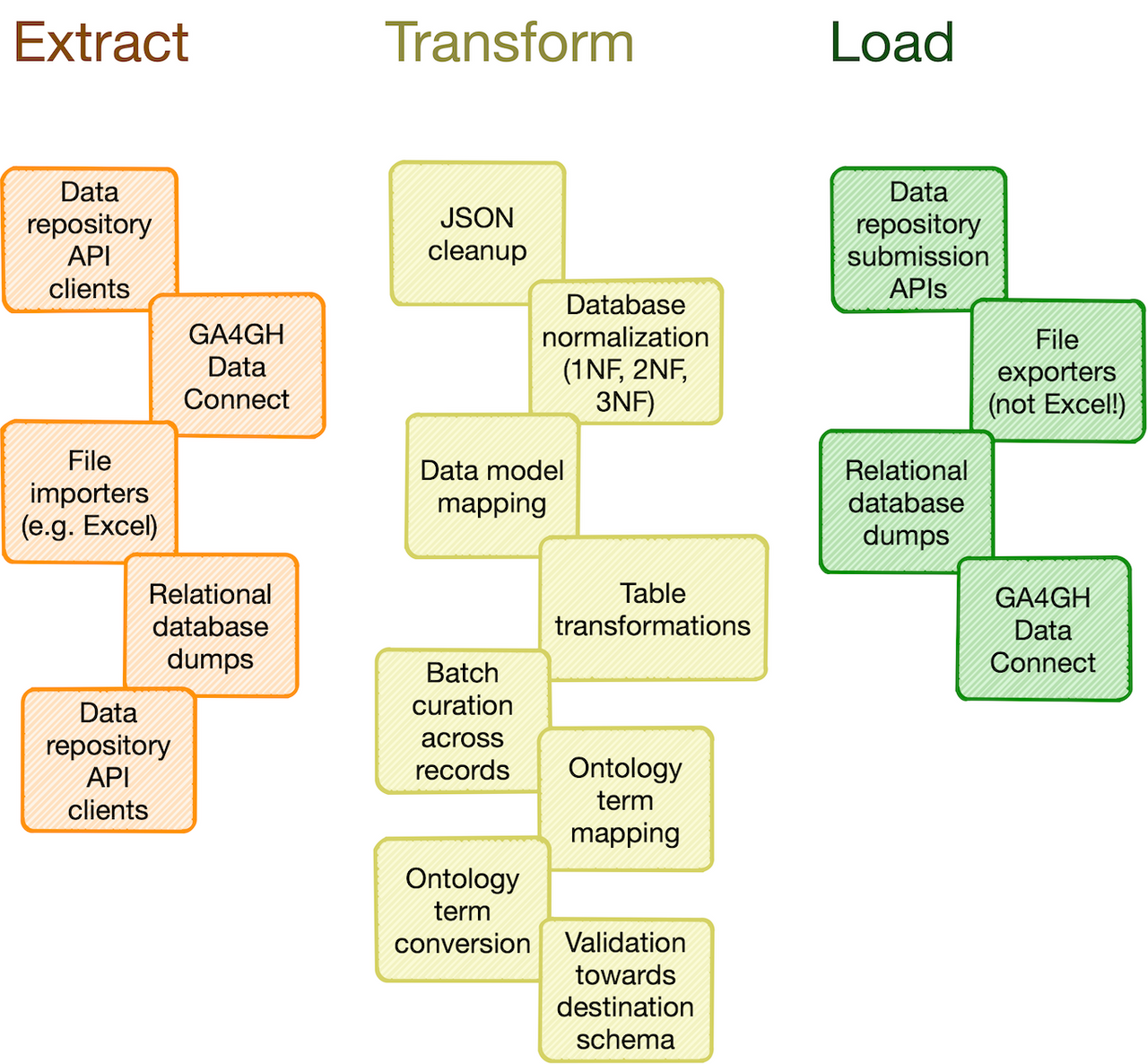
Researchers in life science and other data-centric fields often need to extract, manipulate and integrate data and/or metadata from different sources, such as repositories, databases or flat files. Much research time is spent on trivial and not-so-trivial details of such "data wrangling":
- reformat data structures
- clean up errors
- remove duplicate data
- map and integrate dataset fields
- etc.
General software for data wrangling and analysis, such as Pandas, R or Frictionless, are useful, but researchers still regularly end up with hard-to-reuse scripts, often with manual steps.
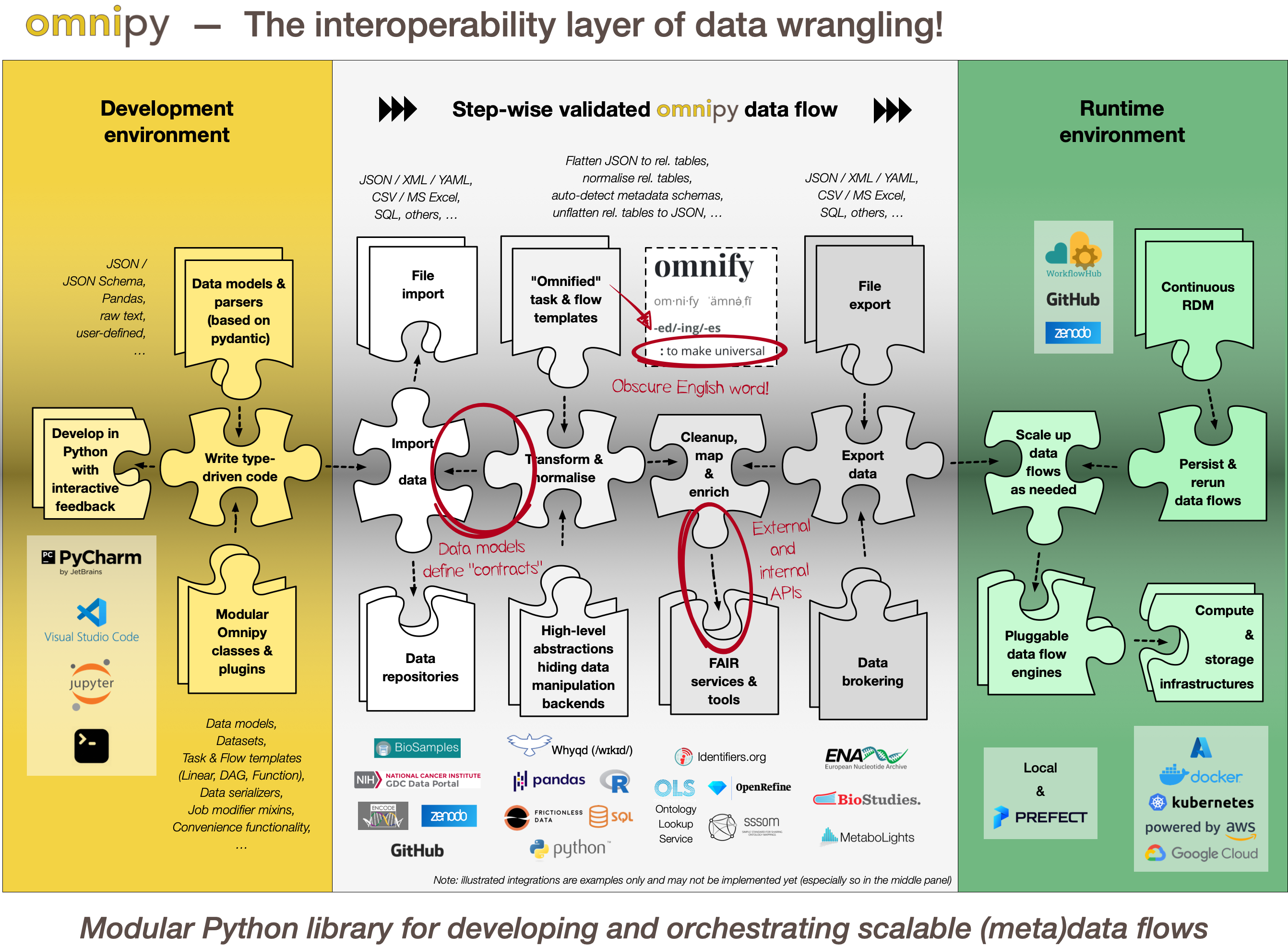
With the Omnipy Python package, researchers can import (meta)data in almost any shape or form: nested JSON; tabular (relational) data; binary streams; or other data structures. Through a step-by-step process, data is continuously parsed and reshaped according to a series of data model transformations.
Omnipy follows the principles of "Type-driven design" (read Technical note #2: "Parse, don't validate" on the FAIRtracks.net website for more info). It makes use of cutting-edge Python type hints and the popular pydantic package to "pour" data into precisely defined data models that can range from very general (e.g. "any kind of JSON data", "any kind of tabular data", etc.) to very specific (e.g. "follow the FAIRtracks JSON Schema for track files with the extra restriction of only allowing BigBED files").
Omnipy tasks (single steps) or flows (workflows) are defined as transformations from specific input data models to specific output data models. pydantic-based parsing guarantees that the input and output data always follows the data models (i.e. data types). Thus, the data models defines "contracts" that simplifies reuse of tasks and flows in a mix-and-match fashion.
Omnipy is built from the ground up to be modular. We aim to provide a catalog of commonly useful functionality ranging from:
- data import from REST API endpoints, common flat file formats, database dumps, etc.
- flattening of complex, nested JSON structures
- standardization of relational tabular data (i.e. removing redundancy)
- mapping of tabular data between schemas
- lookup and mapping of ontology terms
- semi-automatic data cleaning (through e.g. Open Refine)
- support for common data manipulation software and libraries, such as Pandas, R, Frictionless, etc.
In particular, we will provide a FAIRtracks module that contains data models and processing steps to transform metadata to follow the FAIRtracks standard.
An Omnipy module typically consists of a set of generic task and flow templates with related data models, (de)serializers, and utility functions. The user can then pick task and flow templates from this extensible, modular catalog, further refine them in the context of a custom, use case-specific flow, and apply them to the desired compute engine to carry out the transformations needed to wrangle data into the required shape.
When piecing together a custom flow in Omnipy, the user has persistent access to the state of the data at every step of the process. Persistent intermediate data allows for caching of tasks based on the input data and parameters. Hence, if the input data and parameters of a task does not change between runs, the task is not rerun. This is particularly useful for importing from REST API endpoints, as a flow can be continuously rerun without taxing the remote server; data import will only carried out in the initial iteration or when the REST API signals that the data has changed.
In the case of large datasets, the researcher can set up a flow based on a representative sample of the full dataset, in a size that is suited for running locally on, say, a laptop. Once the flow has produced the correct output on the sample data, the operation can be seamlessly scaled up to the full dataset and sent off in software containers to run on external compute resources, using e.g. Kubernetes. Such offloaded flows can be easily monitored using a web GUI.
Offloading of flows to external compute resources is provided by the integration of Omnipy with a workflow engine based on the Prefect Python package. Prefect is an industry-leading platform for dataflow automation and orchestration that brings a series of powerful features to Omnipy:
- Predefined integrations with a range of compute infrastructure solutions
- Predefined integration with various services to support extraction, transformation, and loading (ETL) of data and metadata
- Code as workflow ("If Python can write it, Prefect can run it")
- Dynamic workflows: no predefined Direct Acyclic Graphs (DAGs) needed!
- Command line and web GUI-based visibility and control of jobs
- Trigger jobs from external events such as GitHub commits, file uploads, etc.
- Define continuously running workflows that still respond to external events
- Run tasks concurrently through support for asynchronous tasks
It is also possible to integrate Omnipy with other workflow backends by implementing new workflow engine plugins. This is relatively easy to do, as the core architecture of Omnipy allows the user to easily switch the workflow engine at runtime. Omnipy supports both traditional DAG-based and the more avant garde code-based definition of flows. Two workflow engines are currently supported: local and prefect.