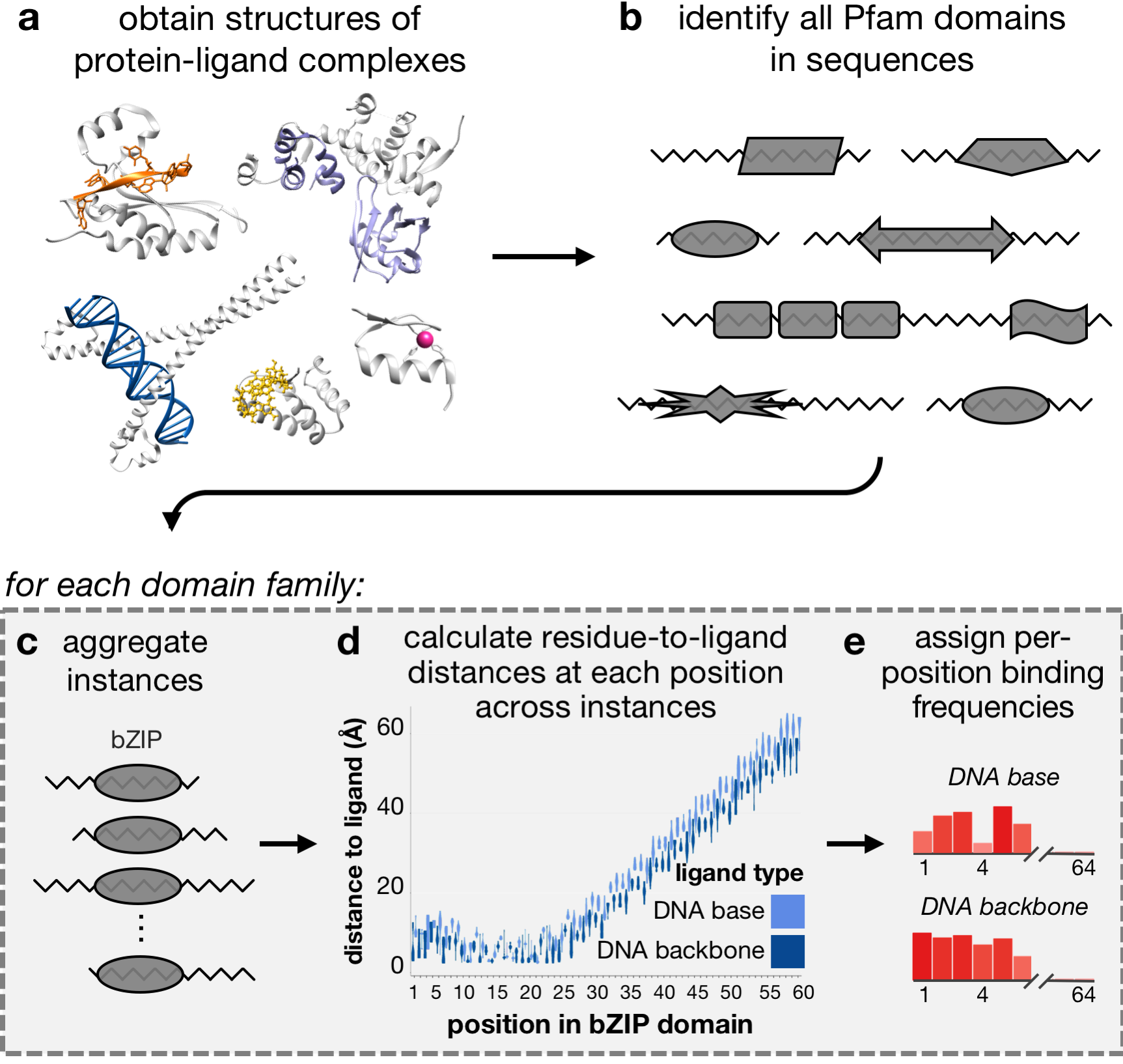
In this project, we use information from protein co-complex structures to determine how individual positions within domains may be involved in binding different ligands. If you use data or scripts from this repository, please cite:
Kobren, S.N. and Singh, M. (2018) "Systematic domain-based aggregation of protein structures highlights DNA-, RNA-, and other ligand-binding positions." Nucleic Acids Res, 47(2): 582–593. doi: 10.1093/nar/gky1224.
InteracDome Web Server
-
To download the primary set of BioLiP data, released March 6, 2013, run the following. Note that this step can take a long time and will require substantial hard drive space (4.1GB).
python download_biolip.py --initialize
-
You must update the BioLiP data that you just downloaded (as BioLiP releases weekly updates) by running:
python download_biolip.py
-
To calculate pairwise Euclidean distances between receptor residue atoms and ligand atoms, run:
python calculate_distances.py --prefix XX
NOTE: This step also takes a long time; we suggest running in parallel. The --prefix option allows you to specify a subset of PDB IDs that begin with a specific 2-character prefix (e.g., 2m).
-
To get the set of all possible prefixes, run:
ls downloaded_data/receptor/ -1 | cut -c1-2 | sort -u
NOTE: This step produces files that take up a lot of space. You must save these files if you are interested in computing alternate distance measures. However, they are otherwise only needed for the immediate next step. To save space wherever you are running in parallel, for each prefix, run the following:
time python calculate_distances.py --prefix AB
time python create_fasta.py --prefix AB
rm -rf processed_data/distances/A/AB/*We suggest using a simple and intuitive score (i.e., ''mindist'') to measure the distances between protein side chains and bound ligands.
-
To calculate the distances between each receptor amino acid residue and all corresponding ligand types, run:
python create_fasta.py --prefix XX
NOTE: Alternate ways of measuring the proximity between a protein receptor chain and ligand (which we found to result in highly correlated values and to take a much longer time to run) are described at the bottom of this page.
You can use whatever method you prefer to find domains in your protein receptor chain sequences.
-
To create a single nonredundant FASTA file of protein chain sequences to search for domain instances, run:
python create_fasta.py --hmmer_input
In our paper, we downloaded all Pfam-A (version 31) domains and ran HMMER locally to find domain hits. General code to run this step of the pipeline can be found at https://github.com/Singh-Lab/run-hmmer.
-
To transform the results obtained from this step to be reflective of original protein chain identifiers (rather than nonredundant identifiers), run:
python create_fasta.py --inflate_nonredundant --original_fasta processed_data/annotations/BioLiP_2018-09-12_nonredundant.fa --nr_results_file processed_data/annotations/BioLiP_2018-09-12-domains-pfam_v31-orig.tsv.gz --output_file processed_data/domains/BioLiP_2018-09-12-domains-pfam_v31.tsv.gz -
The output of these steps, run on BioLiP (version 2018-09-12) can be obtained by running:
if [ ! -d processed_data/domains ]; then mkdir processed_data/domains; fi BIOLIP_DOMAINS="BioLiP_2018-09-12-domains-pfam_v31.tsv.gz" wget http://compbio.cs.princeton.edu/interacdome/$BIOLIP_DOMAINS -O processed_data/domains/$BIOLIP_DOMAINS
NOTE: If you choose to run this domain-finding step independently, you must format the results to match the tab-delimited formatting in the file provided (to run subsequent steps of the pipeline).
The "uniqueness" of each domain sequence must be assessed to account for structural redundancies across PDB entries. To do this, we generate multiple sequence alignments for each domain in contact with each type of ligand, and then assign per-sequence scores as in S Henikoff & JG Henikoff (1994).
-
To get per-domain-instance uniqueness weights, run:
python evaluate_uniqueness.py --create_alignments python evaluate_uniqueness.py
-
Finally, we use the per-domain sequence uniqueness evaluations generated in the previous step to assign per-domain-position binding frequencies, for each ligand type:
python generate_domain_scores.py
Next, we compute the 10-fold cross-validated precision at different binding frequency thresholds for each domain-ligand pair.
-
To get the cross-validated precision achieved at each binding frequency, run:
python cross_validate_scores.py --start X --end X
NOTE: This step may take a long time. You have the option of specifying a subset of domains to run on. The minimum allowed start index is 0, and the maximum end value is the total number of domains for which there are binding frequencies.
-
To find the total number of domains to iterate over, run:
ls processed_data/domains/binding_scores/mindist | wc -l
Finally, to recreate the two data files required for the InteracDome webserver, run the following, remembering that you must specify the full path to a directory containing all Pfam HMMs:
python interacdome_webserver.py --pfam_path <path_to_hmms> --webserverNOTE: The following scripts run optional steps.
You can create lists of domains and binding frequencies that pass different cutoffs by running the following:
python interacdome_webserver.py --pfam_path <path_to_hmms>Possible options and their default values (corresponding to the "confident" set of domain-ligand pairs in the InteracDome paper) are listed below:
| Argument | Default | Description |
|---|---|---|
| ‑‑precision | 0.5 | minimum cross-validated precision (where all instances were split into 10 folds) to consider a particular domain-ligand pair |
| ‑‑grouped_precision | 0. | minimum cross-validated precision (where groups of instances with ≥90% sequence identity were split into 10 folds) to consider a particular domain-ligand pair |
| ‑‑threshold_precision | 0.5 | minimum (ungrouped) cross-validated precision achieved for a binding frequency to be used to infer ligand-binding sites |
| ‑‑structures | 3 | minimum number of distinct PDB entries containing a domain-ligand pair instance |
| ‑‑instances | 0 | minimum number of domain-ligand pair instances |
| ‑‑unique_instances | 3 | minimum number of domain-ligand pair instances with nonredundant sequences |
| ‑‑groups | 0 | minimum number of domain-ligand pair instances with <90% sequence identity |
The previous steps automatically group ligands into the ION_, DRUGLIKE_ and METABOLITE_ super groups using molecular information from the Chemical Component Dictionary, DrugBank and the Human Metabolome Database respectively, resulting in the following file (provided in this repository):
downloaded_data/ligand_groups.txtTo repeat these steps to generate an up-to-date version of the file, the following script can be run
(this code likely requires additional configuration). You first need to install the command-line program
babel from http://openbabel.org/wiki/Main_Page. You will also need to manually download a file from DrugBank
(instructions will be printed to screen when you attempt to run the following), but all other required input
files will be automatically downloaded as needed. You must have at least 8GB of space to download all required
files. The downloaded_data/ligand_groups.txt file was last updated on June 11, 2021.
python group_ligand_types.py --parse_raw --database hmdb
python group_ligand_types.py --parse_raw --database drugbank
python group_ligand_types.py --tanimoto --database hmdb --start 1 --end 31256
python group_ligand_types.py --tanimoto --database drugbank --start 1 --end 31256
python group_ligand_types.py --create_group_list --tanimoto_cutoff 0.9Calculating all-against-all pairwise Tanimoto coefficients can take a while, we suggest running this step in parallel on ~100 BioLiP ligands at a time (e.g., --start 1 --end 100, --start 101 --end 200, ...)
There are multiple ways to assign a continuous ligand-specific binding score to each amino acid residue in a protein receptor chain. These scores are summarized in the table below, where "receptor-ligand atom pair" always refers to a heavy (i.e., non-hydrogen) atom from the side chain of the amino acid residue at a particular position in the protein receptor chain, paired with a heavy atom from the ligand.
| Abbreviation | Score Name | Brief Description |
|---|---|---|
| mindist | minimum distance | minimum distance (in Å) across all receptor-ligand atom pairs for each position in the protein receptor chain |
| fracin4 | fraction within 4Å | fraction of heavy, side-chain atoms that are within 4.0Å of the ligand |
| maxstd | maximum overlap area between normals with σ=1.5 | maximum total overlap area (integrated from -∞ to ∞) between each heavy side-chain receptor atom and all heavy ligand atoms, where the two Gaussian distributions φ(μ=0; σ=1.5) and φ(μ=D; σ=1.5) are centered at each atom in the receptor-ligand atom pair, and where D is the Euclidean distance between the two specified atoms |
| meanstd | average " σ=1.5 | as above, but the average (across all heavy side-chain receptor residue atoms) total overlap area |
| maxvdw | maximum " σ=vdw radii | the maximum total overlap area (as in maxstd) but where the standard deviations of each Gaussian distribution in a receptor-ligand atom pair are set to the van der Waals interaction radii of the respective atoms (rather than a uniform 1.5). |
| meanvdw | average " σ=vdw radii | as above, but the average total overlap area |
To use the maxstd, meanstd, maxvdw, or meanvdw binding frequency calculations, you must first compute the overlap area between two partially overlapping Gaussian distances.
-
Update the previously-computed distance files by running:
python calculate_distances.py --update_overlap --prefix XX
-
Then, generate alternate binding frequency scores by running:
python create_fasta.py --distance <abbreviation> --prefix XX python evaluate_uniqueness.py --create_alignments --distance <abbreviation> python evaluate_uniqueness.py --distance <abbreviation> python generate_domain_scores.py --distance <abbreviation>
In our paper, we also present results showing the bootstrapped standard error of all binding frequencies, across domain-ligand pairs, as well as consistencies of domain-to-ligand distances across 50-50 random splits of domain-ligand pair instances.
-
To generate bootstrapped standard errors of binding frequencies, run:
python cross_validate_scores.py --stderr --distance <abbreviation> --start X --end X
-
To generate distance consistencies for each domain-ligand pair, run:
python cross_validate_scores.py --consistency --distance <abbreviation> --start X --end X