-
Notifications
You must be signed in to change notification settings - Fork 0
Home
The KPFM_sim is an python,cython and shell package managing DFT level simulation of nc-AFM or KPFM measurements.
It contains shell scripts for running important task on the super-computer environments or for the post processing.
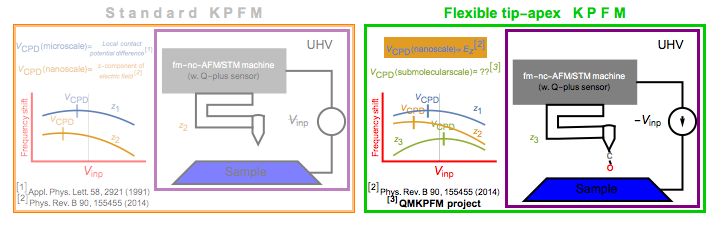
This work is part and one of the main outcome of the QMKPFM project funded by European Union through European Research Committee program Horizon 2020 under project number: 845060. By clicking here or at the flag you will be redirected to a page describing the QMKPFM project.
The DFT part is operated by an open-source CP2K code which is LCAO & PW hybrid with a high amount of options. (!!! Safety warning -- CP2K is an bio-hazard tool -- NEVER, EVER expect proper manual from it, or safe settings for stable, converging calculations. Those do not exist. It has 1000s of options for different types of calculations, but they develop very rapidly and what was working couple of versions ago becomes obsolete quickly. Also the manual is super outdated. Prepare for blood and tears, while working with it. !!! ). However in order to communicate with CP2K via python, the scripts are using a pycp2k package and CP2k_mtools for parsing the output or managing multiple run's.
The (atomic) geometry is handled by the Atomic Simulation Environment -- ASE also together with the previously mentioned suspects -- pycp2k package and CP2k_mtools.
The electric field is calculated through inner C++ procedure calculated in E_field folder
Finally, the results (and the planned task) is kept in an SQL database file (xxx.db), which is handled through an sqlite3.
-
ASE needed. (
python3 setup.py install --user) -
CP2K: CP2K 7.1 working getting .xml file
cp2k.psmp --xml -
cython:
python3 setup.py install --user -
pycp2k: Install through
python3 setup_manual.py install --user; Use the 'xml' file for the setup. ; As for the mpi executable -- use onlysrun; For mahti: insetup.py/install_requiresadding: 'scipy==1.5.3' ; and changing numpy to 'numpy==1.17.5'
- KPFM_sim -- this package
- CP2k_mtools
In the git-cloned KPFM_sim folder navigate to /E_field folder and compile the C++ part through:
make
These two tools are not necessary anymore, but are probably still part of dependencies, which were not cured:
(Once more my appologizes as cython packages are not used in the latest version, but still they are part of the dependencies ... )
python3 setup_cube_import.py build_ext ——inplace
python3 setup_cube_file_writer.py build_ext ——inplace
python3 setup_locpot_import.py build_ext ——inplace
python3 setup_axisym_to_3d_interpolation.py build_ext ——inplace
python3 setup_grid_extparam_interpolation.py build_ext ——inplace
In every example you have to adjust the slurm variables (queue type and so on.) and the PYTHONPATH in the (slurm) ....slrm and (shell) ....sh scripts , so it would match with your settings.
Example with and without the k-points applied in the calculations. To run the full example please run consecutively these comands; if there is a free line wait, until previous run is done without mistakes.
sbatch run_preopt.slrm # optimize the geometry and calculate the initial db file, which is coppied couple of times
sbatch run_afm_worker_1.slrm # calculate every second point of tip descending above Na.
sbatch run_afm_worker_2.slrm # calculate all the other points of tip descending above Na.
sbatch run_afm_worker_y-Cl.slrm # calculate full path of tip descending above Cl (tip is translated).
combine_results.sh # get the results from 3 directories and 3 db files into the global_results directory.
sbatch run_afm_forces.slrm # will rerun CP2K once more, and get the atomic forces from this run. Necessary if you want really high precise atomic forces -- otherwise forces are calculated only on not-fixed atoms (dE forces are normally fine, if the grid is fine enough !! )#
get_results.sh # will create Energy and Force (dE/ds and atomic forces) vs. distance data and plots.
First prepare the system -- the sample and tip model all together. But the tip and sample part has to be separated by the order: tip can be the first and sample the second, or the other way round, we have example scripts for both. Beware the vertical axis is the y axis. So you need to translate your coordinates accordingly (For right angle boxes the transition is as follows: x -> x, y -> z, z -> -y + new-cell-y ). The system in /example is shown here:

Also prepare two files with indices of fixed atoms for the tip and sample parts. (you can either follow the python logic or a human logic, just check and change the pre-optimizing script accordingly).
After this settings adjust the pre-optimizing script. First choose what is first in your atoms order: Tip or Sample and use the right pre-opt. script. Then set the scanning point, and the amount of tip/sample atoms and also the layers: Both tip and sample will be during the run divided by top - middle - bottom. Top and bottom are fixed for tip and sample, respectively. Tip - bottom is one atom tip-apex. Lastly, do not forget about the python/human logic in the numbering of the fixed atoms.
You can check if everything is set properly before running the actual script by setting: just_check = True in the preopt_... scripts. (see either example or control_script_templates ).
If everything runs properly, the script should create a database file with energy, properly divided atom-types.
After the system is pre-optimised. Look at the plan_descend_tip_task.py script and adjust the scanning point and path (stariting, ending point and step) according to what you want. !!! Beware that the original height was set in the pre-optimisation !!!.
The only way, how to make run the descending tip. For the calculations you need to have only the global database <global_results_db_file>. The planning script will create another 2 database files: <task_db_file> and <result_db_file>. The first one will have inside only scan-points (from the global database), while the later one will have inside also the necessary geometries.
If you want to do a different scanning point, than the original geometry (which is normally set to: 0.0 0.0) just adjust x and y accordingly, the translation !!! of the tip !!! is done automatically.
k-points: If you are using k-points, please go here!
To run the calculation run the run_task.py file (ideally in new directory) through slurm. This will consecutively run CP2K calculations and store the results of each of them in the <result_db_file> and the wave-function file. It will take some time.
If the run is interrupted (the handler is currently not working); the resubmitting of the same script is working completely fine - it finds what is already calculated in the <result_db_file> and skip those.
Beware, if the run was interrupted in between storing the wave-function file in the dft stage you need to rm *.wfn or *.kp, you will experience an error and need to remove those files in the worker_*** directory.
k-points: If you are using k-points, please go here!
!!! TO DO: error handler not working yet !!!
After trying the first descend and tune the very moody CP2K to run, you can plan the whole 2D scanning matrix, with z scan in each xy point through creating the whole thing, via create_xy_scan.sh.
This will take a pdtt.template file (pdtt stands for plan_descent_tip_task) and pdtt_run.template (this file should be adjust according to your slurm on your supercomputer) create a whole set of pdtt.x.y.py and rw_x.y.slrm files, each of them can start or restart the whole tip descent for a given xy point.
Each of the rw_x.y.slrm files has to be submitted separately (shell script ...), some more better workflow manager would be much better in the future.
These calculations will create each its own worker folder in the directory, where the calculations are taking space. You can delete those workers after all the z descent is done in each worker.
Also very important - do not forget to merge all the afm_res_X_Y.db files into the main db files, through combine_results.sh
If your calculations are done with the k-points, than the wave-function file (which is kept for later recalculations) has different extension. To work with them, use --kpts while running all the following scripts:
python plan_descend_tip_task.py (options here has no effect on the actual run) -f <task_db_file> <result_db_file> <global_res_db_file>
python run_task.py (optionally -k or --kpts if k-points are necessary; --no_forces if you do not want to calculate the BAD forces; --no_wfn if you don't want to store the wave-function file) -f <task_db_file> <project_path> -s <slurm_id> -t [type_constraint] -c [status_constraint] # the file is now called run_task_w_forces.py
python combine_workers_to_global.py (optionally -k or --kpts if k-points are necessary) <worker1_res_db_file> <worker1_path> <worker2_res_db_file> <worker2_path> <to: global_results_db_file> <global_results_wfn_path>
python copy_scan_points_and_wfn_files.py (optionally -k or --kpts if k-points are necessary) from_db_file <worker_path> to_db_file <results_wfn_path>
python calc_atomic_forces.py (optionally -k or --kpts if k-points are necessary) <project_path> <result_db_file> <global_res_db_file>
While these wavefunction files are important for faster calculations of DFT forces, but from my experience dE/dz works right fine.
The electrostatic force was added to the top through creating an approximate -V/2 on the sample and V/2 on the tip. The field would look like this:

The expected strategy for that would be that the field will charge charge the tip with electrons (attracted by the extra positive potential) and positively charge the sample like this:

This would end up, so the final voltage was supposed to be sample; however the final results do not confirm this, they were rather in the oposite direction, but rather noisy anyway.
NOTE: Our later calculations find out, that 2 metal layers were not enought for the calculations. So we added an extra metal layer, just for the calculations of the electric potential.
The field is calculated to be static and the same, for all the atoms of the sample and all the atoms of the tip, while the potential drop would happen just in the vacuum in between them. The charge is fixed on the shere of 1.0 Angstrom on every atom. The field is then calculated as a solution of the Laplace equation on a final grid for points out of
NOTE: we did not find a big differences of changing the size of atoms which could be done in bias_to_cube_c.py file we generally suggest to read this file, for providing more parameters, that are necessary!
Calculation of the electric field can be done on multiple cores for different geometry on each core (no easy way, how to MPI calculation of a single geometry).
This could by running create_V_field.sh from slurm_script_templates in the computation directory which is using mpicE.template and mpicE_run.template, which should be copied to the same directory.
The calculations of the electric field could be now started through submitting created mpicE_XX.sh scripts.
The calculated field will be store in *.cube files in a separate folder of E_field in your directory.
At the latest version the calculation of the KPFM calculations are done with fixed geometry, and pre-calculated electry field for the initial charge transfer.
Once the electric field is calculated you can create the files for the DFT calculations for different voltages (normally 0.1 V step between -2.0 and +2.0 V) through running create_V_scan.sh script in your calculations directory, which is using pdtt_V_run.template and pdtt_V.template for creating the python and slrm script that creates pdtt_x_y_V.py and rw_x_y_V.slrm files, where the second one can be submitted.
This will run the first calculation which provided electric field, saves the wavefunction and after that calculated 1scf step from the wavefunction, without the extra electric field, thus using the opposite field.
After combining the data into the main database file, you can get the force(x,y,V) infromation through extract_descend_tip_data.py python script. The extracted data were then fitted to parabolas in Wolfram Mathematica, but the data were pretty noisy.
