Infer non-local structural dependencies in genomic sequences. Genomic sequences are esentially compressed encodings of phenotypic information. This package provides a novel set of tools to extract long-range structural dependencies in genotypic data that define the phenotypic outcomes. The key capabilities implemented here are as follows:
- Compute the Quasinet (Q-net) given a database of nucleic acid sequences. The Q-net is a family of conditional inference trees that capture the predictability of each nucleotide position given the rest of the genome. The constructed Q-net for COVID-19 and Influenza A H1N1 HA 2008-9 is shown below.
| COVID-19 | INFLUENZA |
|---|---|
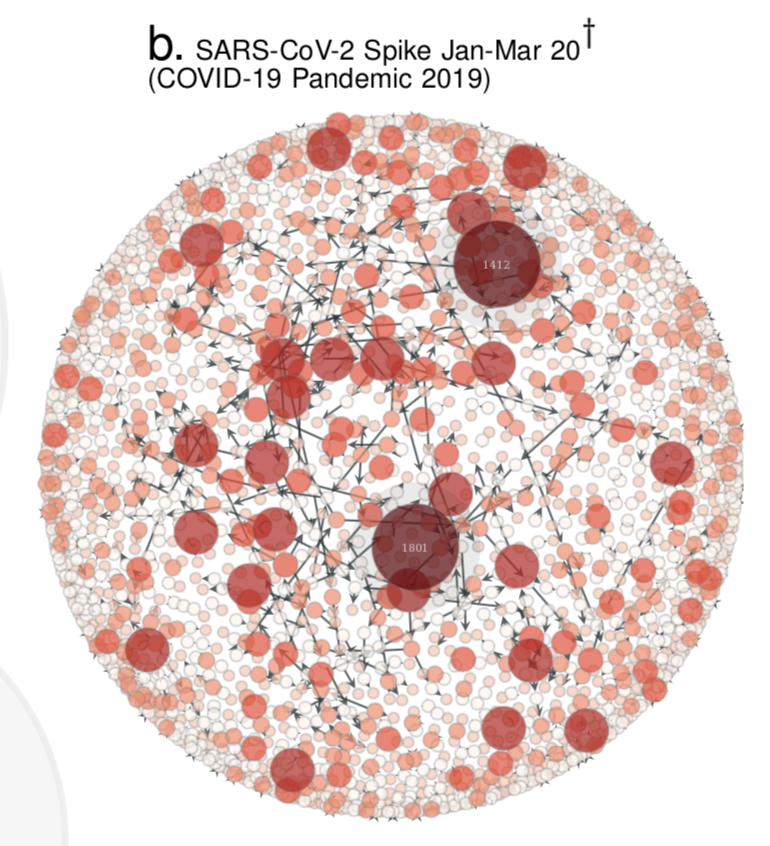 |
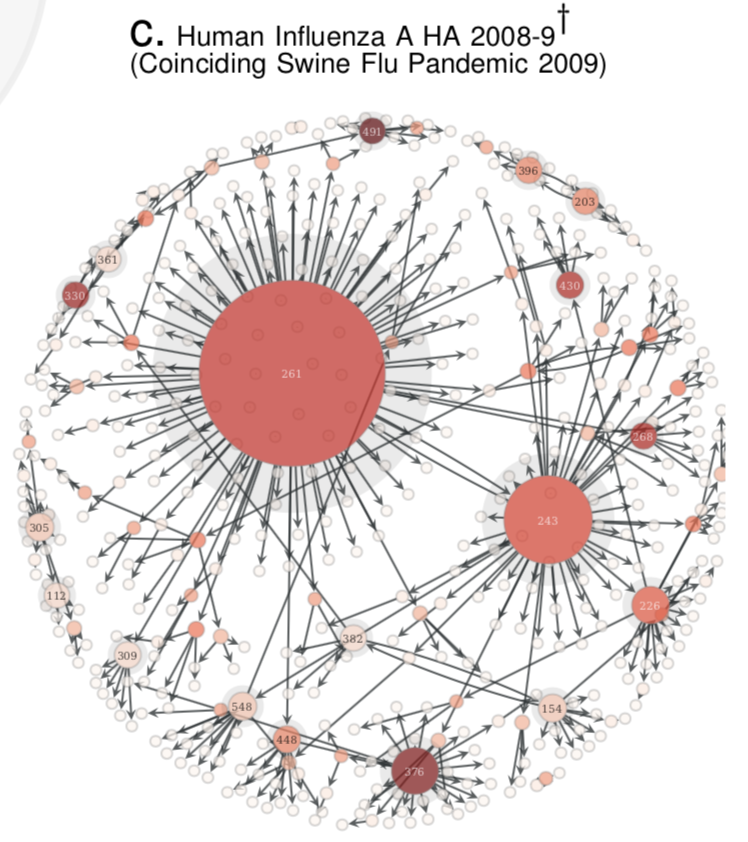 |
-
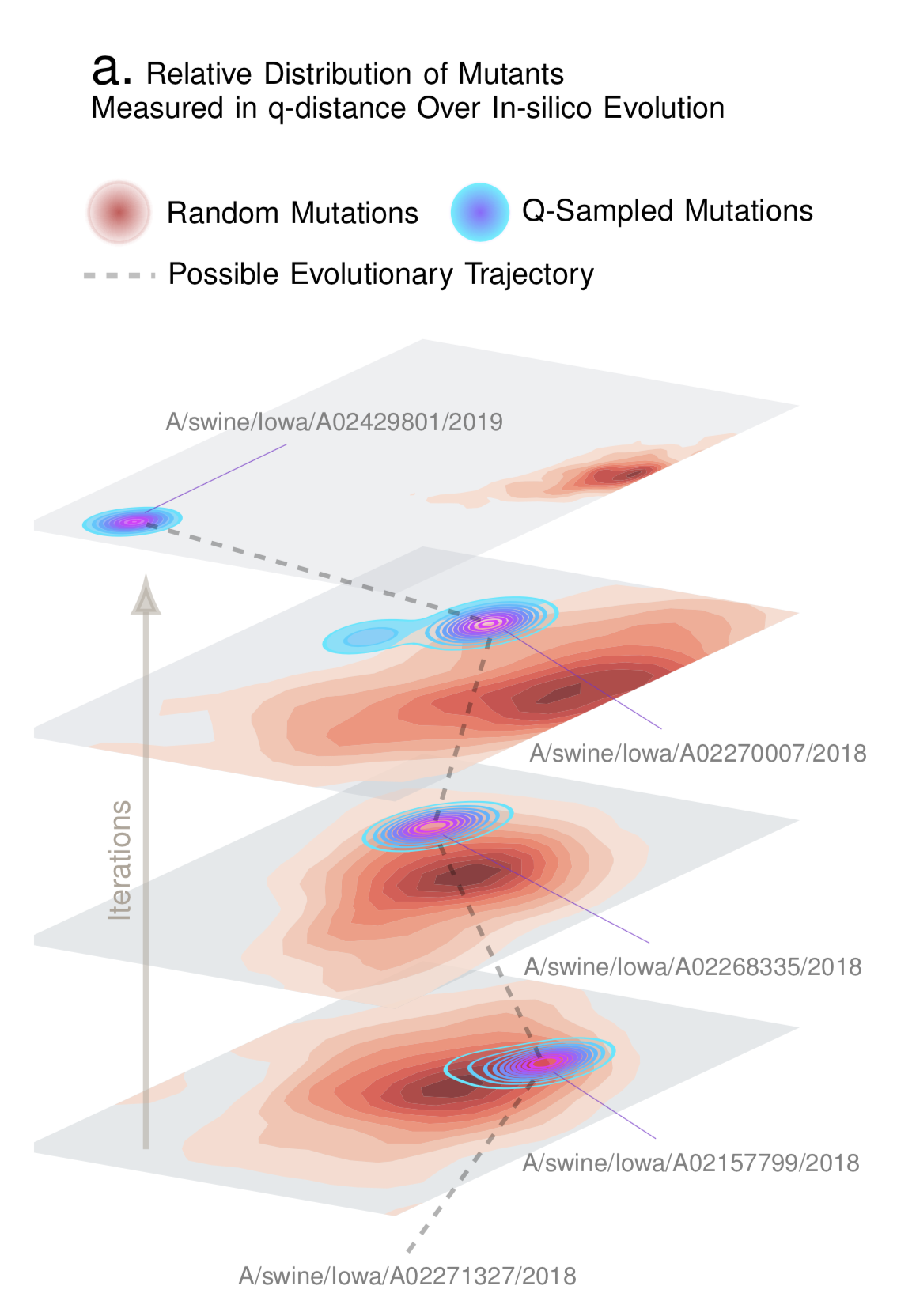
Compute a structure-aware evolution-adaptive notion of distance between genomes, which is demonstrably more biologically relevant compared to the standard edit distance.
-
Draw samples in-silico that have a high probability of being biologically correct. For example, given a database of Influenza sequences, we can generate a new genomic sequence that has a high probability of being a valid influenza sequence.
To install with pip:
pip install quasinet
NOTE: If trying to reproduce the paper below, please use pip install quasinet==0.0.58
- scikit-learn
- scipy
- numpy
- numba
- pandas
- joblib
- biopython
from quasinet import qnet
# initialize qnet
myqnet = qnet.Qnet()
# train the qnet
myqnet.fit(X)
# compute qdistance
qdist = qnet.qdistance(seq1, seq2, myqnet, myqnet)
Examples are located here.
For more documentation, see here.
For reference, please check out our paper:
You can reach the ZED lab at: zed.uchicago.edu