The Python toolkits designed to control the fragment quality of Bulk/SingCell ATAC-seq.
python3 -m pip install --upgrade ATACFragQC
# Basic usage
ATACFragQC [options] -i <input.bam> -r <reference.gtf>
# For more information
ATACFragQC -h
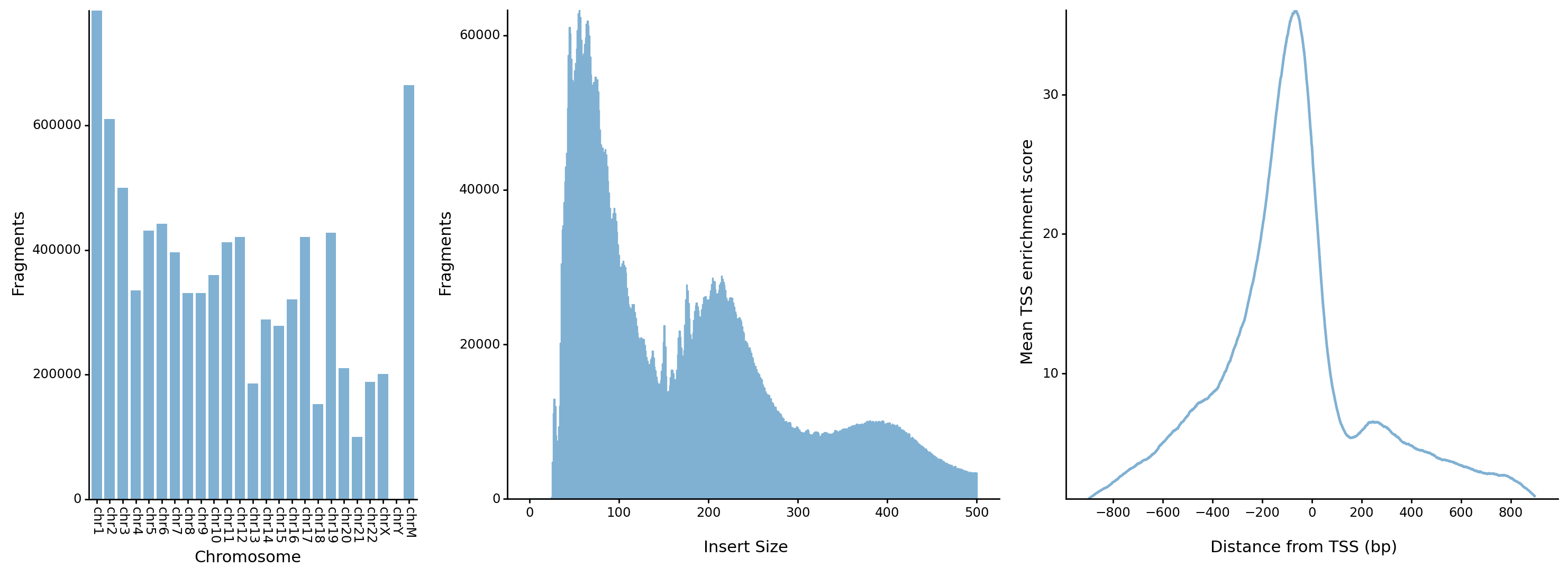
- The distrubution of fragments in chromosomes
- The distrubution of fragment lengths
- The distrubution of fragments around transcription start sites (TSSs)
- Other feature would be supported in the future ...