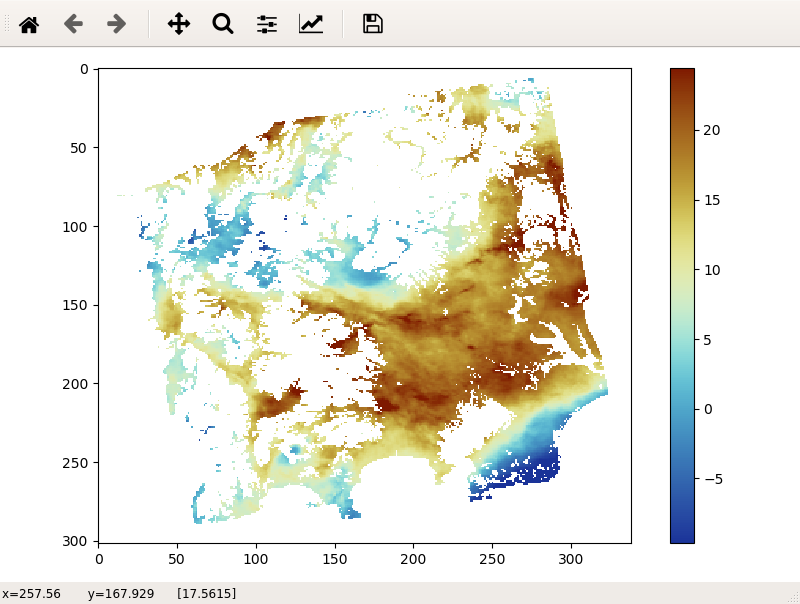
-
Notifications
You must be signed in to change notification settings - Fork 112
3_visualization
For more details of the commands, use -h option to see the usage.
LiCSBAS_disp_img.py -i image_file -p par_file [-c cmap] [--cmin float] [--cmax float] [--auto_crange float] [--cycle float] [--bigendian] [--png pngname] [--kmz kmzname]
-i Input image file in float32 or uint8
-p Parameter file containing width and length (e.g., EQA.dem_par or mli.par)
-c Colormap name (see below for available colormap)
- https://matplotlib.org/tutorials/colors/colormaps.html
- http://www.fabiocrameri.ch/colourmaps.php
- insar
(Default: SCM.roma_r, reverse of SCM.roma)
--cmin|cmax Min|max values of color (Default: None (auto))
--auto_crange % of color range used for automatic determinatin (Default: 99)
--cycle Value*2pi/cycle only if cyclic cmap (i.e., insar or SCM.*O*)
(Default: 3 (6pi/cycle))
--bigendian If input file is in big endian
--png Save png (pdf etc also available) instead of displaying
--kmz Save kmz (need EQA.dem_par for -p option)
This script displays an image file.

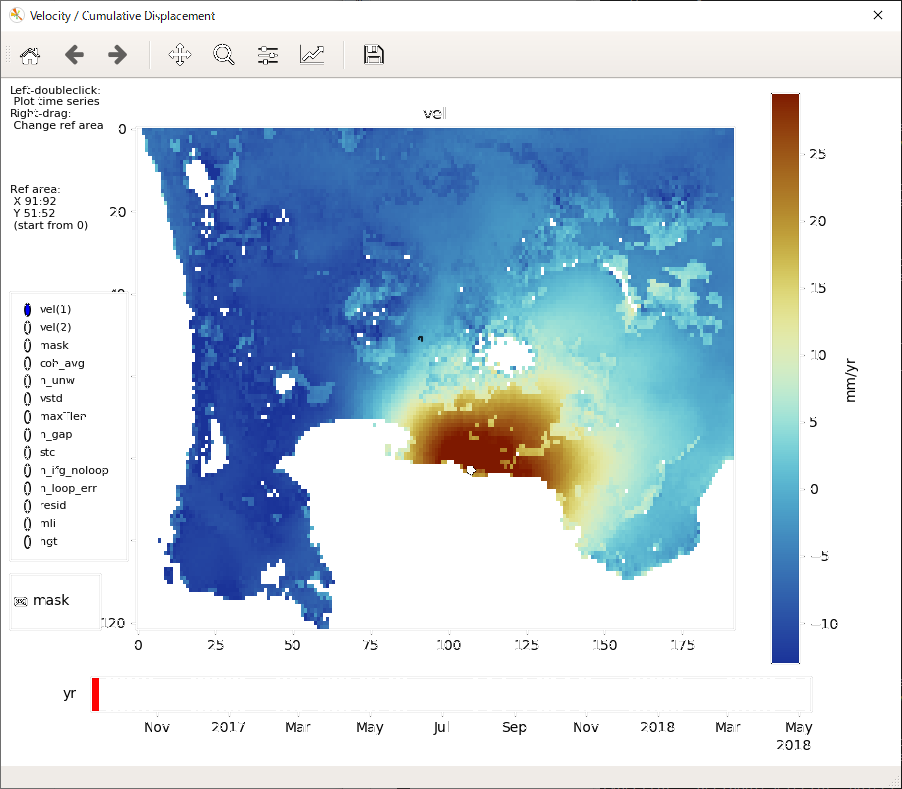
LiCSBAS_plot_ts.py [-i cum[_filt].h5] [--i2 cum*.h5] [-d results_dir] [-m yyyymmdd] [-r x1:x2/y1:y2] [-p x/y] [-c cmap] [--nomask] [--vmin float] [--vmax float] [--auto_crange float] [--dmin float] [--dmax float] [--ylen float]
-i Input cum hdf5 file (Default: ./cum_filt.h5 or ./cum.h5)
--i2 Input 2nd cum hdf5 file
(Default: cum.h5 if -i cum_filt.h5, otherwise none)
-m Master (reference) date for time-seires (Default: first date)
-d Directory containing noise indices (e.g., mask, coh_avg, etc.)
(Default: "results" at the same dir as cum[_filt].h5)
-r Initial reference area (Default: same as info/*ref.txt)
0 for x2/y2 means all. (i.e., 0:0/0:0 means whole area).
-p Initial selected point for time series plot (Default: ref point)
-c Color map for velocity and cumulative displacement
- https://matplotlib.org/tutorials/colors/colormaps.html
- http://www.fabiocrameri.ch/colourmaps.php
(Default: SCM.roma_r, reverse of SCM.roma)
--nomask Not use mask (Default: use mask)
--vmin|vmax Min|max values of color for velocity map (Default: auto)
--dmin|dmax Min|max values of color for cumulative displacement map
(Default: auto)
--auto_crange Percentage of color range used for automatic determinatin
(Default: 99 %)
--ylen Y Length of time series plot in mm (Default: auto)
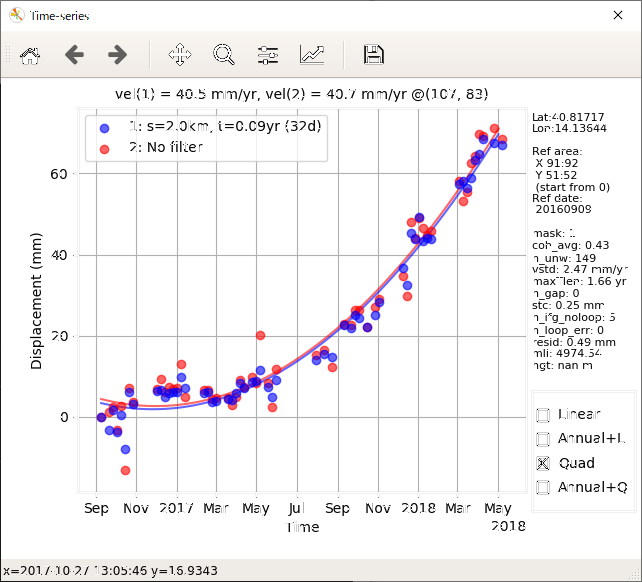
This script displays the velocity, cumulative displacement, and noise indices, and plots the time series of displacement. You can interactively change the displayed image/area and select a point for the time series plot. The reference area can also be changed by right dragging.
Demonstration Video (High resolution mp4, 20MB)
LiCSBAS_profile.py -i infile -p dempar [-r x1,y1/x2,y2] [-g lon1,lat1/lon2,lat2] [-o outfile] [--bigendian] [--nodisplay]
-i Input file (float, little endian)
-p Dem parameter file (EQA.dem_par)
-r Point locations in xy coordinates
-g Point locations in geographical coordinates
-o Output text file (Default: profile.txt)
Format: lat lon x y distance value (x/y start from 0)
--bigendian If input file is in big endian
--nodisplay Not display quick look images
Note: either -r or -g must be specified.
This script gets a profile data between two points specified in geographical coordinates or xy coordinates from a float file. A quick look image is displayed and a text file and kml file are output.

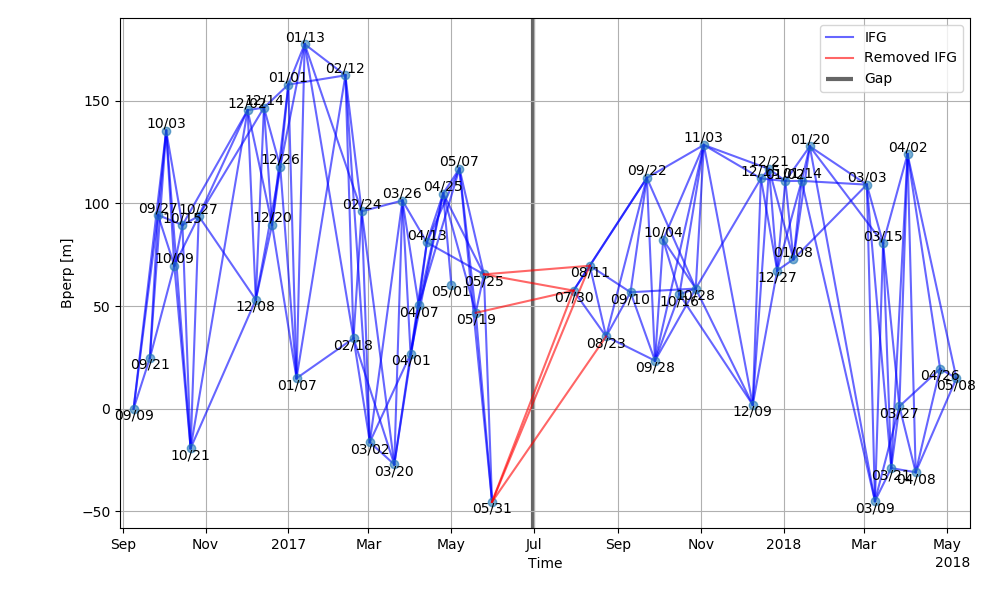
LiCSBAS_plot_network.py -i ifg_list -b bperp_list [-o outpngfile] [-r bad_ifg_list] [--not_plot_bad]
-i Text file of ifg list (format: yyymmdd_yyyymmdd)
-b Text file of bperp list (format: yyyymmdd yyyymmdd bperp dt)
-o Output image file (Default: netowrk.png)
Available file formats: png, ps, pdf, or svg
(see manual for matplotlib.pyplot.savefig)
-r Text file of bad ifg list to be plotted with red lines (format: yyymmdd_yyyymmdd)
--not_plot_bad Not plot bad ifgs with red lines
This script creates a png file (or in other formats) of SB network. A Gap of the network are denoted by a black vertical line if a gap exist. Bad ifgs can be denoted by red lines.