This repository contains Python code to play with SimpleITK and nnU-Net.
- Install Python 3.X,
- Install the required packages:
pip install -r requirements.txt- Download data from the Combined (CT-MR) Healthy Abdominal Organ Segmentation (CHAOS) challenge,
- Extract the archive to
data/.
Alternatively, data can be found in Releases on Github.
python convert_to_nii.py- Follow instructions for the folder structure of data, as detailed for nnU-Net.
- Run the
playing_with_pytorch.ipynbnotebook.
Training was performed with:
- a training dataset of 5 images downsampled to 4x4x4mm,
- no cross-validation (in which case the validation dataset is the same as the training dataset to find the best checkpoint and to decrease the learning rate).
Training for 1 epoch can require 2m30s, but up to 10m, depending on the machine which you obtained.
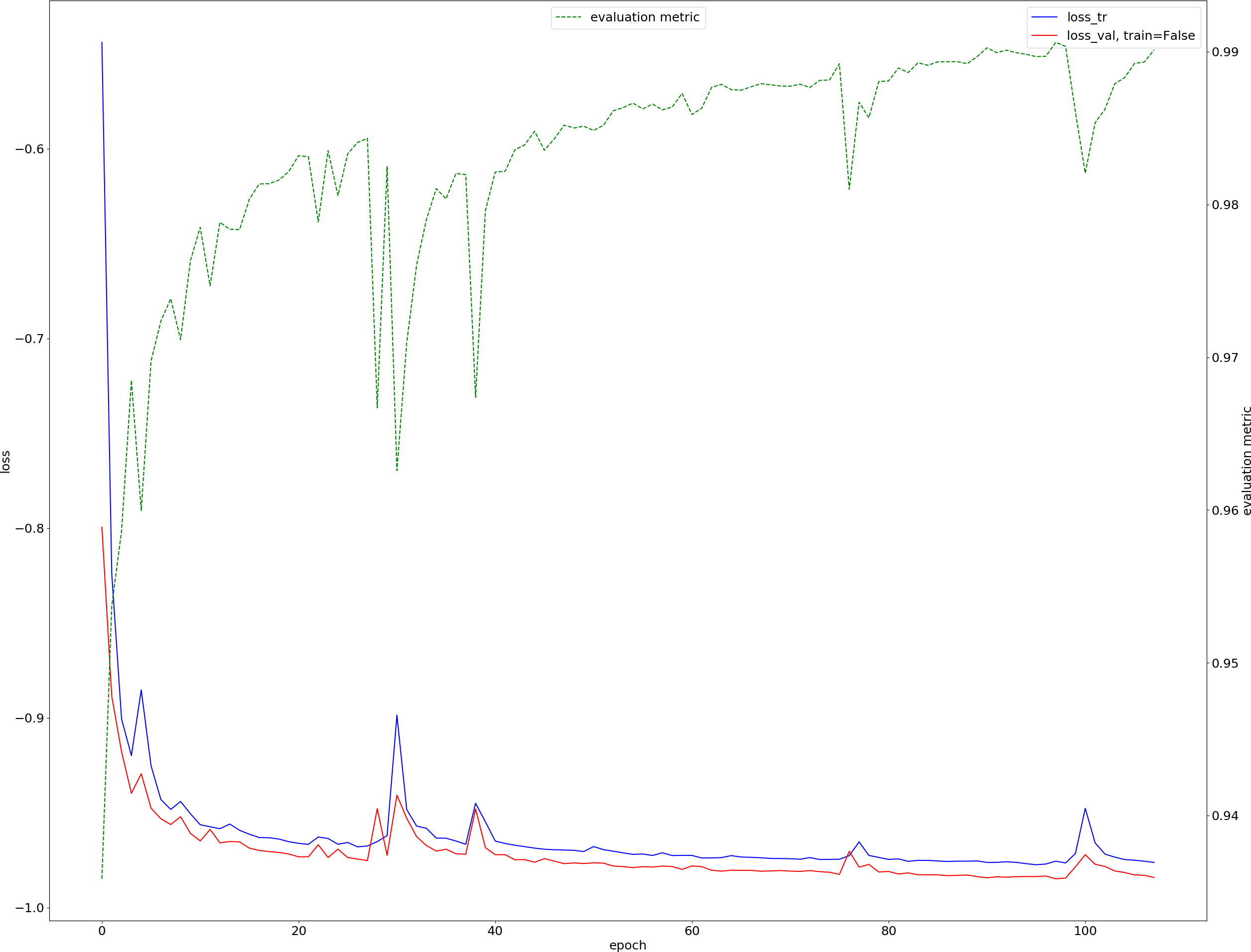
Results obtained after ~ 100 epochs are shown below:
where:
- training loss is in blue,
- validation loss is in red,
- evaluation metric (to maximize) is in green.
In terms of Dice scores, the average is:
- 99.1% for the 5 patients of the training data,
- 94.8% for the 15 patients of the validation data (training data not used).
- Github repository for SimpleITK,
- Notebooks for SimpleITK,
- Documentation for SimpleITK,
- nnU-Net,
- Combined (CT-MR) Healthy Abdominal Organ Segmentation (CHAOS) challenge,
- Evaluation of segmentation results with tools provided by organizers of CHAOS.