-
Notifications
You must be signed in to change notification settings - Fork 0
/
03-analysis.Rmd
411 lines (254 loc) · 13.8 KB
/
03-analysis.Rmd
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
249
250
251
252
253
254
255
256
257
258
259
260
261
262
263
264
265
266
267
268
269
270
271
272
273
274
275
276
277
278
279
280
281
282
283
284
285
286
287
288
289
290
291
292
293
294
295
296
297
298
299
300
301
302
303
304
305
306
307
308
309
310
311
312
313
314
315
316
317
318
319
320
321
322
323
324
325
326
327
328
329
330
331
332
333
334
335
336
337
338
339
340
341
342
343
344
345
346
347
348
349
350
351
352
353
354
355
356
357
358
359
360
361
362
363
364
365
366
367
368
369
370
371
372
373
374
375
376
377
378
379
380
381
382
383
384
385
386
387
388
389
390
391
392
393
394
395
396
397
398
399
400
401
402
403
404
405
406
407
408
409
410
411
# (PART) Analysis {-}
# Data science
Read [Advanced R](https://adv-r.hadley.nz/) and [Efficient R](https://csgillespie.github.io/efficientR/).
Others resources
* [rOpenSci resources](https://ropensci.org/resources/)
* [StackOverflow R tag](https://stackoverflow.com/questions/tagged/r)
* [An aggregated list of free resources](https://github.com/alastairrushworth/free-data-science)
* [Hands-on Scientific Computing](https://handsonscicomp.readthedocs.io/en/latest/)
# Functions
October 25 2018 - Functions in R
[Slides](https://slides.robitalec.ca/functions-in-r.html)
[Resources](https://gitlab.com/robit.a/workshops/-/archive/master/workshops-master.zip?path=functions-in-r)
```{r, echo = FALSE}
knitr::include_url('https://slides.robitalec.ca/functions-in-r.html')
```
# data.table {#data-table}
`data.table` is Alec's favorite R package because it is incredibly efficient,
lightweight and has incredibly responsive and dedicated developers (thank you!). `dplyr` and
etc `tidyverse` packages are full of great functions and workflows, so check those
out as well. Life isn't about strange, strict binaries/camps - use what works
for you.
Note: if you are having install issues on Mac, check out this [reference](https://github.com/Rdatatable/data.table/wiki/Installation#openmp-enabled-compiler-for-mac) can be really useful.
## Syntax
<!-- **TODO: ALR basic resources, and grab from slides** -->

`DT[i, j, by]`
* [List columns](http://brooksandrew.github.io/simpleblog/articles/advanced-data-table/#columns-of-lists) to store a column of lists, or complex objects within a data.table.
* [`.SD`](https://rdatatable.gitlab.io/data.table/articles/datatable-sd-usage.html) = Subset of the Data.table, [.SD vignette](https://rdatatable.gitlab.io/data.table/articles/datatable-sd-usage.html)
* [data.table website](https://rdatatable.gitlab.io/data.table/)
* [Andrew Brooks - Advanced data.table](http://brooksandrew.github.io/simpleblog/articles/advanced-data-table/)
## Slides {#dt-slides}
Workshop: Introduction to data.table
Date: November 16 2017
[Slides](https://slides.robitalec.ca/intro-data-table.html) and [Resources](https://gitlab.com/robit.a/workshops/-/archive/master/workshops-master.zip?path=intro-data-table)
```{r, echo = FALSE}
knitr::include_url('https://slides.robitalec.ca/intro-data-table.html')
```
## Functions
* `fread`/`fwrite` (https://github.com/Rdatatable/data.table/wiki/Convenience-features-of-fread)
* `year`, `month`, `yday`, `mday`, `hour`, `minute`, `second`, `as.IDate`, `as.ITime`
* [`fcase`](https://rdatatable.gitlab.io/data.table/reference/fcase.html): fast case when. When column == value, return x, when column == value2, return x2, etc.
* [`fifelse`](https://rdatatable.gitlab.io/data.table/reference/fifelse.html): fast if-else with sensible behaviour
# targets
`targets` is Alec's other favorite package. It is a tool for combining your
functions and data into a full analysis pipeline. Targets (like predecessors
`drake` and GNU Make), monitors changes to data and R code to only rerun
what you need to, when you need to. It has helped me build huge pipelines
with thousands of datasets, and many analytical steps. I won't attempt to
introduce it here - the package is well documented by a dedicated, and incredibly
helpful developer (thanks!). The pay off here is huge, write functions in R
and easily rerun your analysis with one step: `tar_make()`.
[`targets` manual](https://books.ropensci.org/targets/)
Some projects in the lab that have used `targets` are:
* Movebank summarizer: [`move-book`](https://github.com/robitalec/move-book)
* Preparing animal movement data with [`prepare-locs`](https://github.com/robitalec/prepare-locs)
* [Caribou swimming paper](https://github.com/wildlifeevoeco/CaribouSwimming) (using `drake`)
# Date times
<!-- TODO: anytime -->
## `parsedate`
If you need to set the tz of the **incoming data** - an option is [`parsedate`](https://github.com/gaborcsardi/parsedate).
Similarly flexible than `anytime`, but allows the input tz to
be specified a bit easier. Internally, dates are always stored as UTC, with the
timezone attribute helping to *print* the times in the specified time zone. On
this day, 14:00 NL time = 17:30 UTC and 14:00 BC time = 22:00 UTC.
```{r, eval = FALSE}
library(parsedate)
times <- c("2004-03-01 14:00:00",
"2004/03/01 14:00:33.123456",
"20040301 140033.123456",
"03/01/2004 14:00:33.123456",
"03-01-2004 14:00:33.123456")
parse_date(times, default_tz = 'America/St_Johns')
#> [1] "2004-03-01 17:30:00 UTC" "2004-03-01 17:30:33 UTC"
#> [3] "2004-03-01 17:30:33 UTC" "2004-03-01 17:30:33 UTC"
#> [5] "2004-03-01 17:30:33 UTC"
parse_date(times, default_tz = 'America/Vancouver')
#> [1] "2004-03-01 22:00:00 UTC" "2004-03-01 22:00:33 UTC"
#> [3] "2004-03-01 22:00:33 UTC" "2004-03-01 22:00:33 UTC"
#> [5] "2004-03-01 22:00:33 UTC"
```
A nice, less sensitive to typos, way of setting a tz might be something like this
```{r, eval = FALSE}
grep('Vancouver', OlsonNames(), value = TRUE)
[1] "America/Vancouver"
> grep('Newfoundland', OlsonNames(), value = TRUE)
[1] "Canada/Newfoundland"
> grep('Hawaii', OlsonNames(), value = TRUE)
[1] "US/Hawaii"
```
# Animal Telemetry Data

Alec developed a `targets` workflow for preparing animal relocation data.
The goal is for everyone to 1) start with the same raw data, 2) use the
`prepare-locs` workflow to generate consistent prepared data and 3)
take these data to their own projects. With everyone starting from the same
raw data and using the same preparation workflow, we hope we can reduce some
redundant preparation logic in projects using the same data and facilitate
collaboration. Note, the `prepare-locs` workflow does not necessarily
apply every form of preparation, it aims to be general use allowing users
do their own, more specific preparation afterwards.
Link: https://github.com/robitalec/prepare-locs
## Using `prepare-locs`
The `prepare-locs` uses the `metadata()` function to return a `data.table`
listing file paths, column names, etc. The workflow only prepares datasets
currently available on your local computer - so just make sure the data
you would like to process is present in the specified path.
Note: the expectation is that the user will use the `metadata` workflow
before `prepare-locs`. If the data you are interested in is *not* listed
at [weel.gitlab.io/metadata](https://weel.gitlab.io/metadata/),
contact Alec to document it. If the data is listed, feel free to contribute
changes or clarifications to the metadata.
See more details on the `metadata` project [here](https://weel.gitlab.io/guide/metadata-1.html).
Steps:

1. Clone the [`metadata`](https://gitlab.com/weel/metadata) and the
[`prepare-locs`](https://github.com/robitalec/prepare-locs) workflows.
2. Make sure the data you would like to prepare is placed in the path listed in [`metadata()`](https://github.com/robitalec/prepare-locs#input).
3. Open the `prepare-locs` project.
4. Run `targets::tar_make()`.
# Spatial Analysis
## Packages
These days, the recommendation is to use `sf` (instead of `sp`) and
`terra` (instead of `raster`). The `sf` package is great and should no
doubt be used instead of `sp`. It drops easily into `ggplot2` and other packages.
`terra` may need to be converted to a raster for compatibility with
some packages or functions.
### `sf`

### `terra`

### `raster`
#### Cropping a raster
To crop a raster to the extent of relocations:
```{r, eval = FALSE}
## Packages
library(data.table)
library(raster)
library(rasterVis)
library(ggplot2)
## Load data
# DT is a data.table
load('data/DT.rda')
# DEM is a raster
load('data/dem.rda')
## Plot
# Plot raster and points
gplot(dem) +
geom_tile(aes(fill = value)) +
geom_point(aes(X, Y), data = DT)
## Crop to points
cropped <- crop(dem, DT[, as.matrix(cbind(X, Y))])
gplot(cropped) +
geom_tile(aes(fill = value)) +
geom_point(aes(X, Y), data = DT)
```
If you'd like to buffer the points first, use `sf`.
```{r, eval = FALSE}
## Optionally buffer points first
library(sf)
pts <- st_as_sf(DT, coords = c('X', 'Y'))
buf <- st_buffer(pts, 1e4)
bufCropped <- crop(dem, buf)
gplot(bufCropped) +
geom_tile(aes(fill = value)) +
geom_point(aes(X, Y), data = DT)
```
## Spatial data
Open sources: Open Street Map, Natural Earth, Canadian Government, Provincial Governments.
Examples of downloading and preparing spatial data from these sources in the [study-area-figures](https://gitlab.com/WEEL_grp/study-area-figures) repository (*-prep.R scripts). Packages used for downloading data include: `osmdata`, `rnaturalearth` and `curl`.
Recently from rOpenSci, Dilinie Seimon, Varsha Ujjinni Vijay Kumar highlighted
many collections of open spatial data, from land cover to administrative
boundaries and air pollution to malaria: https://rspatialdata.github.io/,
https://ropensci.org/blog/2021/09/28/rspatialdata/
## Distance to
Calculating distance to something, eg. distance from moose relocations to the
nearest road, using Alec's `distanceto` package.
It can be installed with the following code
```{r, eval = FALSE}
install.packages('distanceto', repos = 'https://robitalec.r-universe.dev')
```
```{r, eval = FALSE}
library(distanceto)
library(sf)
points <- st_read('some-point.gpkg')
lakes <- st_read('nice-lakes.gpkg')
distance_to(points, lakes)
```
Here are the docs: https://robitalec.github.io/distance-to/
# Social Network Analysis
## `spatsoc`
Alec, Quinn and Eric developed `spatsoc`, an R package for generating social
networks from GPS data.
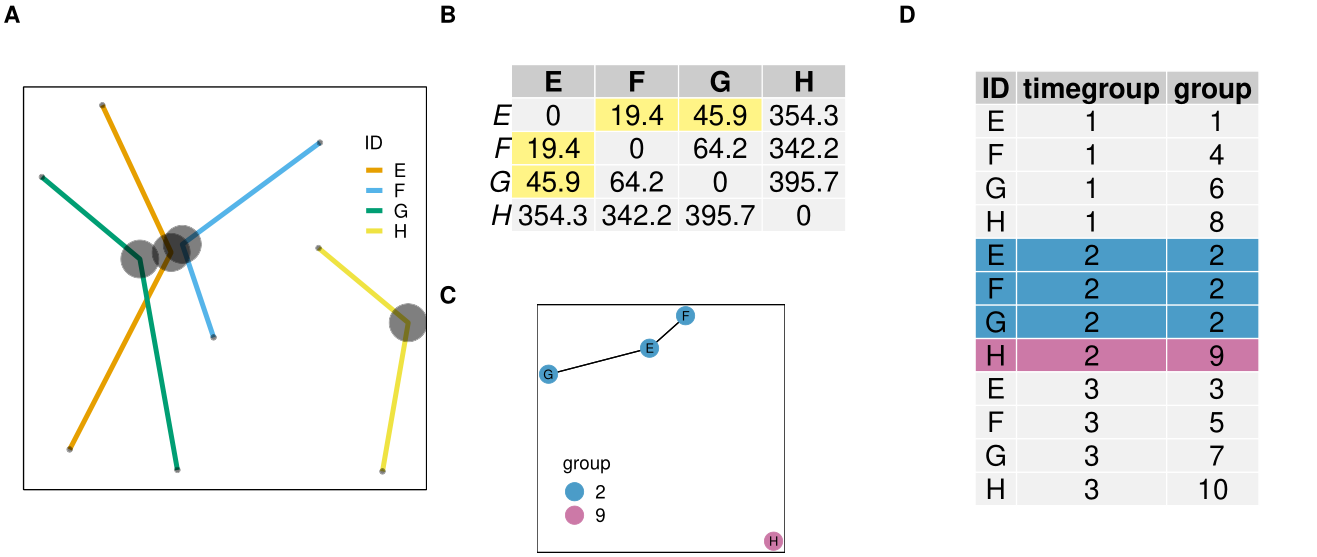
Resources:
* [blog post](https://ropensci.org/blog/2018/12/04/spatsoc/)
* repository: [ropensci/spatsoc](https://github.com/ropensci/spatsoc/)
* vignettes, documentation: [docs.ropensci.org/spatsoc](https://docs.ropensci.org/spatsoc/)
* paper: [Conducting social network analysis with animal telemetry data: Applications and methods using spatsoc](https://besjournals.onlinelibrary.wiley.com/doi/abs/10.1111/2041-210X.13215)
* [CSTWS webinar](https://slides.robitalec.ca/CSTWS-webinar-spatsoc.html)
## Examples
The [Social Network Analysis](https://github.com/wildlifeevoeco/SocCaribou/blob/master/scripts/4-SocialNetworkAnalysis.R)
step in the SocCaribou project is a great place to start.
Alec also recently made a `targets` workflow for `spatsoc`, so feel free to
check that out too: [targets-spatsoc-networks](https://github.com/robitalec/targets-spatsoc-networks)
## `hwig`
Alec developed `hwig`, an R package for calculating gregariousness adjusted half-weight index.
* repository: [robitalec/hwig](https://github.com/robitalec/hwig)
## Slides {#sna-slides}
```{r, echo = FALSE}
knitr::include_url('https://slides.robitalec.ca/CSTWS-webinar-spatsoc.html')
```
<iframe src="https://docs.google.com/presentation/d/e/2PACX-1vRMzDEm8bkxbERxRummBcgqHzOb2vLasEgJNsNC8r462hANEUCwu4dxDyVkdmroFCK7HFsIkhawKe_t/embed?start=false&loop=false&delayms=3000" frameborder="0" width="668" height="396" allowfullscreen="true" mozallowfullscreen="true" webkitallowfullscreen="true"></iframe>
# Integrated Step Selection Analysis

## Slides
<iframe src="https://docs.google.com/presentation/d/e/2PACX-1vTkKdK9fAzU6ARUdJLhd_lhE8CNphNECuDb99k2lWwOOQFqi2a_oNnA1zBTNCWNbw/embed?start=false&loop=false&delayms=3000" frameborder="0" width="668" height="396" allowfullscreen="true" mozallowfullscreen="true" webkitallowfullscreen="true"></iframe>
<iframe src="https://docs.google.com/presentation/d/e/2PACX-1vRLZ5vWK7WAwoedENJ4EyNFQO9vhuhnn9x3mSW6rE6hqtOXZG6Ip0y1aCCjcJsnDw/embed?start=false&loop=false&delayms=3000" frameborder="0" width="668" height="396" allowfullscreen="true" mozallowfullscreen="true" webkitallowfullscreen="true"></iframe>
## Projects
Alec and Julie are working on a `targets` workflow for iSSA.
* https://gitlab.com/robit.a/targets-amt-issa
# Earth Engine
## Resources
<!-- TODO fill best practices, EE group, repo of my scripts-->
[Google Earth Engine Developer guide](https://developers.google.com/earth-engine/guides)
## Slides
Workshop: An Introduction to Remote Sensing with Earth Engine
Authors: Alec L. Robitaille, Isabella C. Richmond
Date: December 10 2020
[Slides](https://slides.robitalec.ca/ee.html)
[Resources](https://github.com/robitalec/workshops/tree/master/ee)
```{r, echo = FALSE}
knitr::include_url('https://slides.robitalec.ca/ee.html')
```
# Behavioural Reaction Norms
## Slides {#brn-slides}
<iframe src="https://docs.google.com/presentation/d/e/2PACX-1vTPmfrBj_WyLSq6AhmTXrAwxGDNzve3F7Dhgwr-01m9yU7WE2tDLttT-18MXrxTyg/embed?start=false&loop=false&delayms=3000" frameborder="0" width="480" height="389" allowfullscreen="true" mozallowfullscreen="true" webkitallowfullscreen="true"></iframe>
# Models

* [`easystats` family](https://github.com/easystats)
## Bayesian methods
Richard McElreath's Statistical Rethinking course has
20 online lectures and 10 weeks of homework and solutions available [here](https://github.com/rmcelreath/statrethinking_winter2019).
Alec's progress through this material is available at:
https://www.statistical-rethinking.robitalec.ca/
# Cool packages {#analysis-cool}

<!-- **TODO: organize into spatial/ plotting/ helper/ etc** -->
## Data frames / tables / sheets
* [`googlesheets4`](https://googlesheets4.tidyverse.org/)
## Dates and times
* [`anytime`](https://github.com/eddelbuettel/anytime)
* [`parsedate`](https://github.com/gaborcsardi/parsedate)
## Spatial packages
* [`parzer`](https://github.com/ropensci/parzer) parsing geographic coordinates