In biomedical research, lymphoblastoid cell lines (LCLs), often established by in vitro infection of resting B cells with Epstein-Barr virus, are commonly used as surrogates for peripheral blood lymphocytes. Genomic and transcriptomic information on LCLs has been used to study the impact of genetic variation on gene expression in humans. Here we present single-cell RNA sequencing (scRNA-seq) data on GM12878 and GM18502—two LCLs derived from the blood of female donors of European and African ancestry, respectively. Cells from three samples (the two LCLs and a 1:1 mixture of the two) were prepared separately using a 10x Genomics Chromium Controller and deeply sequenced. The final dataset contained 7,045 cells from GM12878, 5,189 from GM18502, and 5,820 from the mixture, offering valuable information on single-cell gene expression in highly homogenous cell populations. This dataset is a suitable reference for population differentiation in gene expression at the single-cell level. Data from the mixture provide additional valuable information facilitating the development of statistical methods for data normalization and batch effect correction.
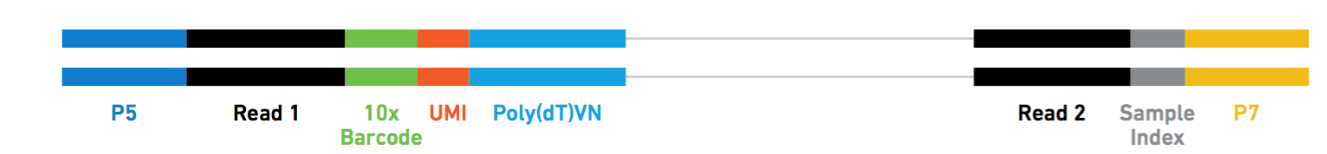 R1 (10X Barcode + UMI + Read1) and R2 (Sample Index + Read2) files (required to run Salmon alevin and Kallisto BUS) are provided through SRA [here]. If you are interested in the 3 fastq files required to run CellRanger, they are available [here]
R1 (10X Barcode + UMI + Read1) and R2 (Sample Index + Read2) files (required to run Salmon alevin and Kallisto BUS) are provided through SRA [here]. If you are interested in the 3 fastq files required to run CellRanger, they are available [here]