A TKinter based python GUI for visualizing Tristan-MP plots. A work in progress.
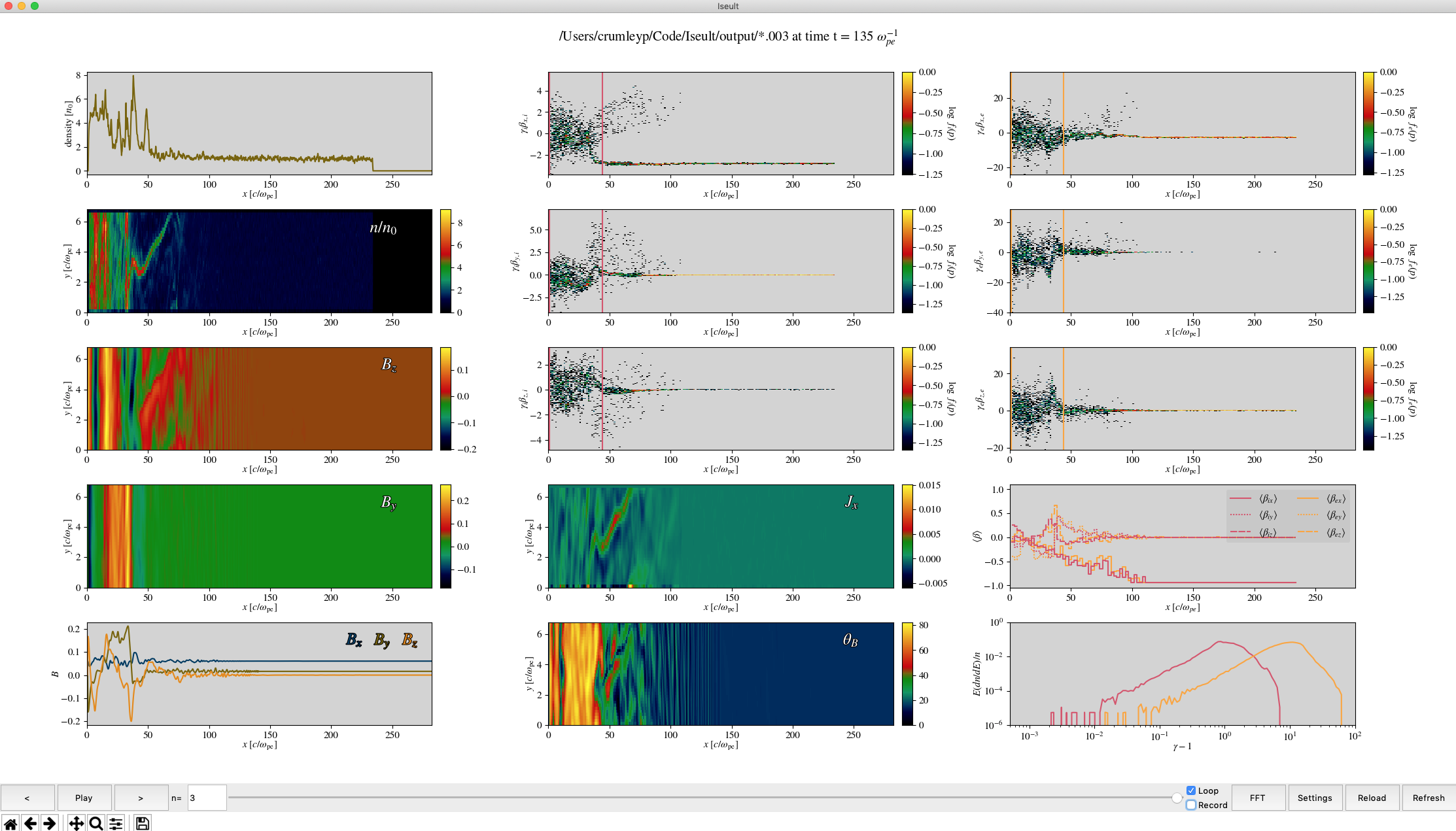 An example visualization of a Tristan-MP simulation.
An example visualization of a Tristan-MP simulation.
Written by:
- Patrick Crumley, patrick.crumley@gmail.com, based on Jaehong's Tristan analysis IDL script.
- Robert Caddy, rcaddy@princeton.edu
July 2024: Ported to Python 3.11, see environment.yml for details on all dependencies. Added support for Tristan v2 data.
May 8th 2019: Ported code to python 3.7.x & Matplotlib 3.0.x There may be a few bugs here and there, but I think it is working.
The code is now it's beta phase. See the implemented column for what has already been implemented.
Python packages required: See the environment.yml file
To use the movie saving feature: ffmpeg & xterm.
Iseult should work on Windows, MacOS & Linux.
There are two different methods to setup what you need for Iseult to run. The recommended method is to setup a python environment with conda, pyenv, etc and a conda environment.yml file is provided. The other method for those with access to the Stellar cluster at Princeton is to simply load the proper Anaconda module.
# Recommended setup
# If on Stellar
$ module load anaconda3/2024.6 # or the latest version
# If not on Stellar then install/load Anaconda
$ cd /path/to/Iseult/
$ conda env create
$ conda activate iseult
$ chmod +x ./iseult.py
# After initial setup
# make sure the proper environment is activated
$ conda activate iseult
# Then run iseult. This method opens a GUI to allow you to search for the data you want to open
$ cd /path/to/Iseult/
$ ./iseult.py
# Or if you want to open data directly
$ cd /directory/that/contains/data # usually the `output` directory
$ /path/to/iseult.py # you can append an ampersand (&) to open iseult in the backgroundThe previous method is still recommended on Stellar, this secondary method is simplier but potentially less reliable if the software stack on Stellar is changed.
# Setup
$ module load anaconda3/2023.3
$ cd /path/to/Iseult/
$ chmod +x ./iseult.py
# After initial setup
$ module load anaconda3/2023.3
# Then run iseult. This method opens a GUI to allow you to search for the data you want to open
$ cd /path/to/Iseult/
$ ./iseult.py
# Or if you want to open data directly
$ cd /directory/that/contains/data # usually the `output` directory
$ /path/to/iseult.py # you can append an ampersand (&) to open iseult in the backgroundWhen Iseult is started, it checks to see if Tristan-MP data is located at the current path, if it isn't Iseult prompts you to select the directory of where your Tristan-MP data is saved. To edit/change any of the plots, just right click on the subplot directly. You can also change the number of columns, the colormap, and other general settings by clicking the settings button. The measure button allows you to take measurements like T_i, T_e, measure spectra, take 1-D FFTs, etc. The matplotlib interactive toolbar is beneath the playbar, it allows you to save the figure, use your mouse to zoom around, etc.
If you get a set-up you like, you can go to file menu and choose Save Iseult State. It will give you an option to name the 'view.' To replace the default state of Iseult, name the view Default. The views are saved as .cfg files in .iseult_configs folder. You must restart Iseult to see the saved config in the preset views menu.
Enjoy!
| Implemented: |
|---|
| Time stepping |
| Movie (without recording) |
| Basic plotting |
| ability to modularly change plots. |
| plot control panels to edit things about indv. plots |
| shock-finding |
| figure saving |
| Ability to take measurements |
| ability to save Iseult settings in a config file |
| zooming |
| Left to Implement: |
|---|
| gifs/movies |
| Longer term goals (???) |
| Useful links |
|---|
| http://python.org |
| http://effbot.org/tkinterbook/ |
| http://matplotlib.org |
| http://matplotlib.org/users/navigation_toolbar.html |
| http://h5py.org |